I´ve run a GLMM model with the following variables: Response variable: continuous (with positive, negative and zero values; Explanatory variable: 1 factor with 6 levels (DB, DF, NDB, NDF, BB, BF); individual as a random effect variable
After running a lmer function, and checking the assumptions, I want to perform a posteriori specific comparisons. How do I do that? I want to test for example if there is any difference between DF vs. DB. How do I write this in R??
CODE IS AS FOLLOWS
#Model:
m1 <- lmer(Vueltasmin ~ Condicion + (1 | Bicho), Datos)
summary(m1)
#Checking asusmptions: OK
#Comparisons:
model.matrix.gls <- function(object, ...) {
model.matrix(terms(object), data = getData(object), ...)
}
model.frame.gls <- function(object, ...) {
model.frame(formula(object), data = getData(object), ...)
}
terms.gls <- function(object, ...) {
terms(model.frame(object), ...)
}
#Comparisons desired:
#DB-DF
#NDB-NDF
#BB-BF
#DB-BB
Here I show how the data plot looks like, and the comparisons desired.
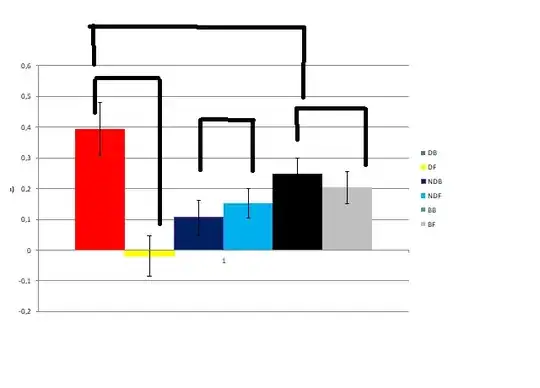
Thanks everyone for the help provided!