Suppose I have some sample data $x_i$ then I can estimate the quantile $Q_p(x_i)$ using for example the quantile() function in R.
Now suppose I add some random noise to the data: $y_i=x_i+\epsilon_i$ (keeping the $x_i$ unchanged) where $\epsilon_i$ are i.i.d. and drawn from some distribution with zero mean.
Is there anything I can say about the distribution of $Q_p(y_i)$?
I've done some numerical experiments in R in which the $x_i$ are constructed at the outset from a normal distribution and then the $\epsilon_i$ are randomly drawn from a known distribution (either uniform or normal). $Q_p(y_i)$ is calculated 500 times with different random $\epsilon_i$ to estimate its distribution.
It looks like $Q_p(y_i)$ follows a bell shaped curve with a larger mean than $Q_p(x_i)$. Is there any theory on this?
R code below:
x <- rnorm(1e6,0,1/qnorm(0.95))
Q_simulated <- rep(NA,500)
for(s in 1:500)
{
epsilon <- rnorm(length(x),0,0.05)
y <- x+epsilon
Q_simulated[s] <- quantile(y,0.95)
}
ggplot(data.frame(x=Q_simulated),aes(x)) + geom_histogram() + geom_vline(xintercept=quantile(x,0.95),colour="red")
EDIT:
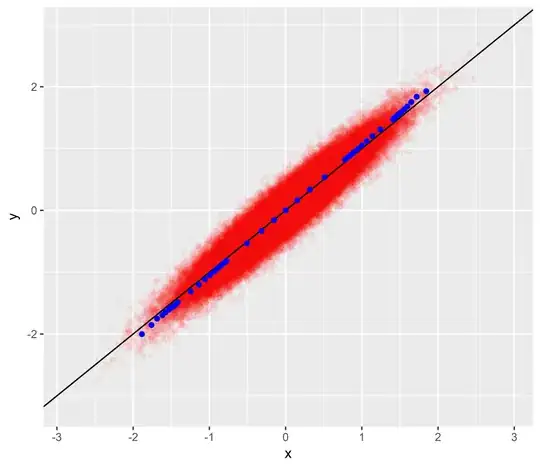
Drawing a scatter plot of $x$ versus $y=x+\epsilon$ in red, superimposing the unit line in black and various quantiles $(Q_p(x),Q_p(y))$ in blue gives the following plot:
x <- rnorm(1e5, 0, 1/qnorm(0.95))
epsilon <- rnorm(length(x), 0, 0.2)
y <- x + epsilon
p <- seq(0.1, 0.9, 0.1)
p <- c(0.01*p, 0.1*p, p, 0.9+0.1*p, 0.99+0.01*p)
qx <- quantile(x,p)
qy <- quantile(y,p)
ggplot(data.frame(x=x, y=x+epsilon), aes(x, y)) + geom_point(colour="red", alpha=0.04) + geom_abline(slope=1, intercept=0) + geom_point(data=data.frame(x=qx, y=qy), colour="blue")