Rajkomar, A., Oren, E., Chen, K., Dai, A.M., Hajaj, N., Liu, P.J., Liu, X., Sun, M., Sundberg, P., Yee, H. and Zhang, K., 2018. Scalable and accurate deep learning for electronic health records. arXiv preprint arXiv:1801.07860. https://arxiv.org/pdf/1801.07860.pdf
Model Evaluation and Statistical Analysis Patients were randomly split into development (80%), validation (10%) and test (10%) sets. Model accuracy is reported on the test set, and 1000 bootstrapped samples were used to calculate 95% confidence intervals. To prevent overfitting, the test set remained unused (and hidden) until final evaluation.
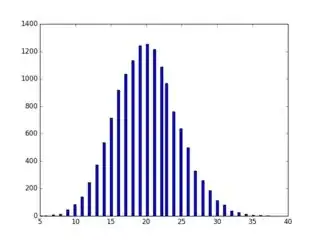
The study involved only 216,221 hospitalizations from 114,003 patients, so taking a single test set, especially with some highly imbalanced outcomes, seems to not make sense. Would not nested CV or bootstrapping the full set have been vastly superior, especially given that some of the diagnoses they were predicting (14,000 different ones) only had perhaps (guessing) 200 cases in the whole dataset ($p$ = 0.001). I think the test set is 20,000.
obs = []
for i in range(0,14000): obs.append(np.random.binomial(n=20000,p=0.001))
A hold out might have only caught 20 positive cases for some diseases (assuming most are rare). Further, bootstrapping the held out test set does not really help to detect this variation.
Generally, for their predictive models, I imagine Google uses a single hold out because their numbers are so large, but for these rare diseases this might be a real issue, and many diseases are rare.
Edit: published fast track https://www.nature.com/articles/s41746-018-0029-1