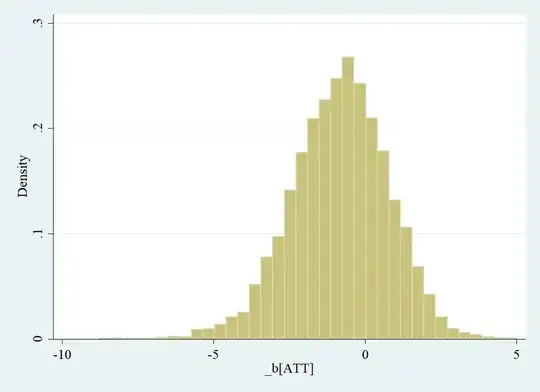
I am using propensity score matching with kernel matching to test for the average treatment effect on the treated.
The distribution of the bootstrapped differences is as follows:
I transformed them with the mean (as described in this CV -answer) and the SD to obtain z-scores. Afterwards, I counted how many observations (in abs. values) exceeded the 95% threshold of 1.65.
However, with this approach I get a p-value of 0.09.
How does this work with a (as in my case) skewed distribution?
If I simply count the observations exceeding the 2.5% and 97.5% seperately, then I obtain a summed p-value of 0.04 which is in line with the BCa confidence interval. Would this be a valid alternative?