I'm fitting poisson GLMM, and I'm quite confused about the need or not to log10 transform my main predictor.
Raw values of this main predictor was very spread, from 2e+03 to 6e+06, that's why I thought about log10 transformation. Linearity with response seem to me equal.
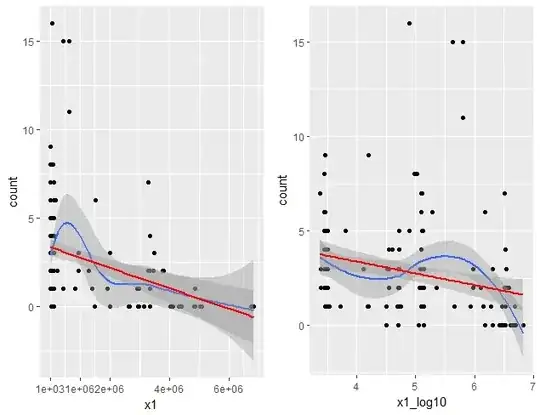
For fitting GLMM I had to scale the predictors (errors without scaling), using:
pvars <- c("x1","x1_log10", "x2" ,"x3", "x4", "x5")
mydf_sc <- mydf
mydf_sc[pvars] <- lapply(mydf[pvars],scale)
Plot with the scaled predictor are :
I'm very confused because results of my GLMM are opposite : my main predictor is significant without log10 transform and not significant if I use log10 transform
glmm1 <- glmer(count ~ x1+ x2 + x3 + x4 + x5 +
(1| x6) +(1|x7)+(1|ID),
data=mydf_sc, family="poisson")
summary(glmm1)
Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) ['glmerMod']
Family: poisson ( log )
Formula: count ~ x1 + x2 + x3 + x4 + x5 + (1 | x6) + (1 | x7) + (1 | ID)
Data: mydf_sc
AIC BIC logLik deviance df.resid
610.8 638.6 -296.4 592.8 152
Scaled residuals:
Min 1Q Median 3Q Max
-1.9743 -0.6970 -0.2632 0.5131 3.0054
Random effects:
Groups Name Variance Std.Dev.
ID (Intercept) 0.07861 0.2804
x7 (Intercept) 0.03236 0.1799
x6 (Intercept) 0.78608 0.8866
Number of obs: 161, groups: ID, 161; x7, 8; x6, 2
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.41893 0.64230 2.209 0.0272 *
x1 -0.49491 0.12024 -4.116 3.86e-05 ***
x2 -0.13887 0.11129 -1.248 0.2121
x3 0.07619 0.09702 0.785 0.4323
x4 -0.08049 0.06327 -1.272 0.2033
x5 -0.09930 0.07945 -1.250 0.2113
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) x1 x2 x3 x4
x1 0.079
x2 -0.034 -0.519
x3 0.041 -0.257 0.514
x4 -0.053 -0.152 -0.003 -0.085
x5 -0.092 -0.125 0.117 0.256 0.297
And with the log10 transform and scaled predictor
glmm2 <- glmer(count ~ x1_log10+ x2 + x3 + x4 + x5 +
(1| x6) +(1|x7) + (1|ID),
data=mydf_sc, family="poisson")
summary(glmm2)
Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) ['glmerMod']
Family: poisson ( log )
Formula: count ~ x1_log10 + x2 + x3 + x4 + x5 + (1 | x6) + (1 | x7) +
(1 | ID)
Data: mydf_sc
AIC BIC logLik deviance df.resid
628.4 656.2 -305.2 610.4 152
Scaled residuals:
Min 1Q Median 3Q Max
-2.0486 -0.6626 -0.1504 0.4169 2.3551
Random effects:
Groups Name Variance Std.Dev.
ID (Intercept) 0.11584 0.3403
x7 (Intercept) 0.03584 0.1893
x6 (Intercept) 0.82438 0.9080
Number of obs: 161, groups: ID, 161; x7, 8; x6, 2
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.50363 0.65939 2.280 0.0226 *
x1_log10 -0.16203 0.13867 -1.168 0.2426
x2 -0.31247 0.13154 -2.376 0.0175 *
x3 -0.05047 0.10111 -0.499 0.6176
x4 -0.12361 0.06499 -1.902 0.0572 .
x5 -0.12676 0.08173 -1.551 0.1209
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) x1_l10 x2 x3 x4
x1_log10 0.090
x2 -0.048 -0.663
x3 0.089 0.176 0.223
x4 -0.035 -0.002 -0.086 -0.116
x5 -0.082 -0.014 0.047 0.219 0.285
If I compare fits with AIC, glmm1 is better (i.e lower) , and if I calculate the sum of square residuals glmm1 is better (ie.lower) too.
I thought to use a log10 transformation because of the spread of the predictor values, but finally since I use scaled predictors, I wonder if it's necessary yet.
So, if some of you can explain me what happens (why results are so different) and which analysis is the good one, it would be very very appreciated.
Data are here :
mydf <- structure(list(count = c(1, 1, 1, 5, 15, 11, 9, 8, 7, 1, 5, 16,
6, 2, 8, 15, 4, 3, 1, 0, 4, 1, 2, 2, 2, 1, 3, 1, 5, 3, 3, 4,
3, 2, 1, 0, 2, 2, 6, 2, 0, 0, 3, 1, 2, 2, 2, 1, 3, 5, 7, 7, 7,
6, 2, 3, 3, 4, 1, 2, 3, 1, 2, 3, 1, 1, 1, 1, 1, 2, 2, 5, 2, 2,
6, 2, 2, 2, 2, 2, 3, 2, 0, 0, 0, 0, 0, 0, 2, 3, 2, 2, 1, 0, 0,
3, 1, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 2, 0, 1,
1, 4, 1, 6, 3, 5, 1, 3, 4, 6, 7, 6, 3, 2, 3, 3, 5, 6, 8, 9, 4,
3, 2, 1, 6, 2, 2, 1, 1, 3, 5, 3, 2, 3, 3, 2, 3, 1, 4, 1, 2, 3,
1, 3, 1), x1 = c(454276.630324255, 15803.1563972592, 15458.2342654783,
79089.1163309219, 433064.92842954, 639609.580040433, 15796.6139883664,
104607.240566262, 3301847.85530658, 3380.36483734805, 6357.74361426188,
78110.710827558, 1529337.73525669, 3474601.85370647, 94724.1554098659,
639609.580040433, 39834.5777550968, 49961.5621483385, 49501.3804401392,
50826.3757249488, 51670.4355390994, 55337.9747884692, 52492.3355531823,
51375.6168345031, 51830.7997135719, 54004.1327091058, 52364.8333586487,
54076.335684573, 52105.8109404304, 52453.8631578501, 35511.3686511835,
35456.7012643244, 33395.0533851741, 35062.9690293352, 31354.2541181611,
31831.853724259, 118596.374688501, 121554.512420281, 191138.31164019,
121100.531704515, 113179.847358967, 137020.588002108, 137085.296834259,
136367.64719088, 136367.64719088, 135610.442532084, 136824.220830818,
136110.128893872, 133403.823145702, 132311.491140916, 128584.592590665,
123079.910041864, 123796.075203802, 124141.510674517, 121886.481343848,
122145.003101152, 13077.9129382755, 124419.09895087, 124419.09895087,
124419.09895087, 124515.585953799, 124515.585953799, 124515.585953799,
124611.257457142, 124611.257457142, 124611.257457142, 124611.257457142,
124419.09895087, 127248.25326102, 127248.25326102, 127248.25326102,
127248.25326102, 127248.25326102, 127248.25326102, 125084.715383792,
116820.543248463, 3312347.83977499, 3307143.68368415, 3339420.73710133,
3339420.73710133, 3489612.02613466, 3787340.40364162, 4044735.09967731,
4332712.49030506, 4410506.3486271, 6738481.68768351, 6829376.07553111,
6753771.27992383, 950841.73646546, 950841.73646546, 230393.74295532,
1283593.72888636, 1419207.9736855, 1491344.05744556, 2013224.87745932,
2023866.97925484, 1925108.17089723, 2661178.20766687, 2922632.22932389,
2972397.52352174, 2973263.36236786, 5087084.6439317, 5062249.54053654,
5049109.16912577, 4874011.01990889, 4865212.37320984, 4844194.80198645,
2946546.02832311, 2646007.37429602, 2678211.41076352, 2018903.43065148,
4123476.19271286, 3164645.53052, 3824227.28626133, 3342110.58530565,
3339420.73710133, 3342110.58530565, 3343192.06281568, 852591.942449119,
2887.67136368804, 2887.67136368804, 2887.67136368804, 5225.19886143861,
2841.08844859385, 2841.08844859385, 2838.0416631723, 2384.70089496048,
2818.29878593123, 2816.21191647018, 2816.21191647018, 2816.21191647018,
2835.9401746766, 2838.0416631723, 2838.0416631723, 2841.08844859385,
2880.08521424055, 2880.08521424055, 2882.21941509514, 2882.21941509514,
2924.40544679865, 2924.40544679865, 3226.70820676332, 3226.70820676332,
3226.70820676332, 3226.70820676332, 3226.70820676332, 3214.82585949069,
3209.8220949141, 2441.3578929725, 2468.63429708923, 2439.58170286854,
2441.3578929725, 2441.3578929725, 3207.28767252863, 3207.28767252863,
3209.77492390452, 3209.77492390452, 3209.77492390452, 3209.77492390452,
3226.70820676332, 3226.70820676332), x1_log10 = c(5.6573203956694,
4.19874383815735, 4.18915988463051, 4.89811672316093, 5.63655301400862,
5.80591495996403, 4.19856400570374, 5.01956174599534, 6.51875705768416,
3.52896357551324, 3.80330301034245, 4.89271059001513, 6.18450340454985,
6.54090504701398, 4.97646074165915, 5.80591495996403, 4.60026021802215,
4.69863600900148, 4.69461731023005, 4.70608914258146, 4.71324212230491,
4.74302326116043, 4.72009589635918, 4.7107570492143, 4.71458790975622,
4.73242699582451, 4.71903972581057, 4.73300725530994, 4.71688615935781,
4.71977747892335, 4.55036741085983, 4.54969832829418, 4.52368214206241,
4.54484868809727, 4.49629647386532, 4.50286193044554, 5.0740714135068,
5.08477108562158, 5.28134774548676, 5.08314604996246, 5.05376910388449,
5.13678582689321, 5.13699087677218, 5.1347113474973, 5.1347113474973,
5.13229313318477, 5.13616298365657, 5.13389044525503, 5.12516827596201,
5.12159756393332, 5.10918893317819, 5.09018717015312, 5.09270687615138,
5.09391702599875, 5.08595553988135, 5.08687570487486, 4.11653844186966,
5.09488705183641, 5.09488705183641, 5.09488705183641, 5.09522371665322,
5.09522371665322, 5.09522371665322, 5.09555727852436, 5.09555727852436,
5.09555727852436, 5.09555727852436, 5.09488705183641, 5.10465182948183,
5.10465182948183, 5.10465182948183, 5.10465182948183, 5.10465182948183,
5.10465182948183, 5.09720424470521, 5.06751922150016, 6.52013593709916,
6.51945306388122, 6.5236711397163, 6.5236711397163, 6.54277714493193,
6.57833434097488, 6.60689008379363, 6.63675987112144, 6.64448845155199,
6.82856205247856, 6.83438102881605, 6.82954634893355, 5.97810823649622,
5.97810823649622, 5.36247068032029, 6.10842758664355, 6.15204604253689,
6.17357784802218, 6.30389228834369, 6.30618196465291, 6.28445513732707,
6.42507395836007, 6.46577416916377, 6.47310689079747, 6.47323337935542,
6.70646896391141, 6.70434354963434, 6.70321476087896, 6.68788650676914,
6.68710180256944, 6.68522159931963, 6.46931322964654, 6.42259105021112,
6.42784485606065, 6.30511554601734, 6.61526349146535, 6.500325072091,
6.58254369590421, 6.52402081593243, 6.5236711397163, 6.52402081593243,
6.52416132706371, 5.93074122396392, 3.46054776609357, 3.46054776609357,
3.46054776609357, 3.7181028235452, 3.45348475436296, 3.45348475436296,
3.45301876672153, 3.3774339146909, 3.44998703355778, 3.44966533182334,
3.44966533182334, 3.44966533182334, 3.45269706498709, 3.45301876672153,
3.45301876672153, 3.45348475436296, 3.45940533759498, 3.45940533759498,
3.45972703932942, 3.45972703932942, 3.46603758412046, 3.46603758412046,
3.50875969365855, 3.50875969365855, 3.50875969365855, 3.50875969365855,
3.50875969365855, 3.50715745305789, 3.50648096220605, 3.38763144985953,
3.39245675841327, 3.38731536744143, 3.38763144985953, 3.38763144985953,
3.50613791502314, 3.50613791502314, 3.50647457983995, 3.50647457983995,
3.50647457983995, 3.50647457983995, 3.50875969365855, 3.50875969365855
), x2 = c(1615L, 1500L, 1530L, 1605L, 1300L, 1367L, 1700L, 1450L,
1550L, 1315L, 1375L, 1455L, 1515L, 1585L, 1650L, 1700L, 900L,
910L, 915L, 920L, 925L, 935L, 990L, 995L, 1000L, 1005L, 1010L,
1015L, 1020L, 1025L, 1030L, 1035L, 1040L, 1045L, 1050L, 1055L,
1175L, 1180L, 1185L, 1190L, 1195L, 1200L, 1205L, 1210L, 1215L,
1220L, 1225L, 1230L, 1235L, 1240L, 1245L, 1250L, 1255L, 1260L,
1265L, 1270L, 1295L, 1300L, 1305L, 1310L, 1315L, 1320L, 1325L,
1330L, 1335L, 1360L, 1365L, 1370L, 1375L, 1380L, 1385L, 1390L,
1395L, 1400L, 1405L, 1410L, 1500L, 1502L, 1505L, 1508L, 1510L,
1512L, 1514L, 1516L, 1518L, 1520L, 1522L, 1524L, 1528L, 1530L,
1532L, 1534L, 1538L, 1540L, 1542L, 1544L, 1546L, 1548L, 1550L,
1552L, 1556L, 1559L, 1602L, 1604L, 1608L, 1612L, 1615L, 1620L,
1633L, 1636L, 1638L, 1640L, 1643L, 1645L, 1648L, 1650L, 1652L,
1654L, 1658L, 810L, 815L, 820L, 825L, 830L, 835L, 840L, 845L,
850L, 855L, 900L, 905L, 910L, 915L, 920L, 925L, 930L, 935L, 940L,
945L, 950L, 955L, 950L, 955L, 1000L, 1005L, 1010L, 1015L, 1020L,
1025L, 1030L, 1035L, 1040L, 1045L, 1050L, 1055L, 1100L, 1105L,
1110L, 1115L, 1130L, 1135L), x3 = c(13.5, 13.5, 13.5, 24, 24,
24, 24, 24, 24, 0, 2, 1, 1, 1, 1, 1, 26, 26, 26, 26, 26, 26,
26, 26, 26, 26, 26, 26, 26, 26, 26, 26, 26, 26, 26, 26, 28, 28,
28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28,
28, 28, 29, 29, 29, 29, 29, 29, 29, 29, 29, 29, 29, 29, 29, 29,
29, 29, 29, 29, 29, 29, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30,
30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30,
30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30,
30, 50, 50, 50, 50, 50, 50, 50, 50, 50, 50, 50, 50, 50, 50, 50,
50, 50, 50, 50, 50, 50, 50, 52, 52, 52, 52, 52, 52, 52, 52, 52,
52, 52, 52, 52, 52, 52, 52, 52, 52, 52, 52), x4 = c(30L, 60L,
30L, 40L, 40L, 20L, 50L, 20L, 10L, 30L, 5L, 25L, 10L, 0L, 15L,
20L, 60L, 60L, 60L, 90L, 20L, 20L, 5L, 20L, 30L, 20L, 30L, 20L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 30L, 5L, 20L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 30L, 40L,
40L, 40L, 40L, 40L, 40L, 40L, 40L, 40L, 40L, 40L, 40L, 5L, 5L,
0L, 0L, 30L, 30L, 40L, 50L, 50L, 40L, 30L, 0L, 0L, 0L, 0L, 20L,
20L, 20L, 0L, 0L, 0L, 0L, 0L, 15L, 15L, 5L, 10L, 10L, 10L, 30L,
50L, 50L, 50L, 50L, 50L, 50L, 50L, 20L, 20L, 20L, 20L, 20L, 20L,
20L, 20L, 20L, 40L, 40L, 40L, 40L, 40L, 40L, 40L, 40L, 0L, 0L,
0L, 30L, 30L, 30L, 10L, 10L, 10L, 50L, 50L, 50L, 50L, 50L, 50L,
40L, 40L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 50L, 50L, 50L, 50L,
50L, 50L, 50L, 50L, 50L, 50L, 0L, 0L), x5 = c(40L, 40L, 70L,
60L, 60L, 70L, 50L, 70L, 50L, 70L, 95L, 50L, 90L, 70L, 80L, 70L,
0L, 0L, 0L, 0L, 10L, 20L, 20L, 10L, 40L, 70L, 50L, 60L, 90L,
90L, 90L, 90L, 90L, 90L, 95L, 95L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 40L, 50L, 30L, 5L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 30L,
20L, 30L, 10L, 40L, 20L, 20L, 30L, 30L, 0L, 0L, 0L, 0L, 5L, 5L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 30L, 40L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 10L, 25L, 45L, 60L, 60L, 60L, 20L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 10L, 10L, 10L, 10L, 20L, 20L, 20L, 20L,
20L, 0L, 0L, 0L, 0L, 0L, 0L, 20L, 20L, 50L, 50L, 50L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 10L, 10L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 50L,
50L), x6 = structure(c(1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L), .Label = c("Date1", "Date48", "Date49",
"Date2", "Date3"), class = "factor"), x7 = structure(c(3L, 4L,
4L, 1L, 3L, 2L, 4L, 2L, 6L, 1L, 7L, 1L, 6L, 6L, 2L, 2L, 8L, 8L,
8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L,
8L, 8L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L), .Label =
c("Site4",
"Site6", "Site1", "Site3", "Site7", "Site9", "Site5", "Site10",
"Site11", "Site13", "Site12", "Site2", "Site8"), class = "factor"),
ID = 1:161), .Names = c("count", "x1", "x1_log10", "x2",
"x3", "x4", "x5", "x6", "x7", "ID"), row.names = c(NA, -161L), class =
"data.frame")
Thanks @Florian Hartig , @whuber, and @Elvis for all the element you gave. They were very helpful to understand what happens. As suggested by @Elvis, I fit the model removing the 4 points having count >10 and obtained pvalue = 0.09.
ind <- which(mydf_sc$count >10)
ind
[1] 5 6 12 16
glmm2b <- glmer(count ~ x1_log10+ x2 + x3 + x4 + x5 +
+ (1| x6) +(1|x7) + (1|ID),
+ data=mydf_sc[-ind,], family="poisson")
summary(glmm2b)
Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) ['glmerMod']
Family: poisson ( log )
Formula: count ~ x1_log10 + x2 + x3 + x4 + x5 + (1 | x6) + (1 | x7) +
(1 | ID)
Data: mydf_sc[-ind, ]
AIC BIC logLik deviance df.resid
592.7 620.2 -287.4 574.7 148
Scaled residuals:
Min 1Q Median 3Q Max
-1.8740 -0.7304 -0.1666 0.4929 2.5919
Random effects:
Groups Name Variance Std.Dev.
ID (Intercept) 0.06340 0.2518
x7 (Intercept) 0.06662 0.2581
x6 (Intercept) 0.51231 0.7158
Number of obs: 157, groups: ID, 157; x7, 8; x6, 2
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.25735 0.54202 2.320 0.0204 *
x1_log10 -0.34372 0.20201 -1.702 0.0888 .
x2 -0.18029 0.15799 -1.141 0.2538
x3 0.01162 0.13034 0.089 0.9289
x4 -0.12246 0.06382 -1.919 0.0550 .
x5 -0.08543 0.08204 -1.041 0.2978
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) x1_l10 x2 x3 x4
x1_log10 0.184
x2 -0.135 -0.726
x3 0.099 -0.135 0.217
x4 -0.055 -0.058 -0.092 -0.050
x5 -0.111 -0.085 0.027 0.257 0.327