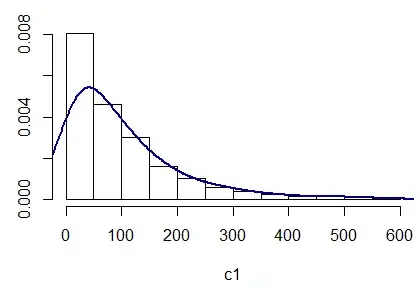
I've got a dataset with similar data "waiting time in seconds", which belongs to an exponential distribution according to graphs. I have tried normal, lognormal as well, but it fits best an exponential distribution.
library(gsheet)
patience.data<-gsheet2tbl('https://docs.google.com/spreadsheets/d/1YIKOiA_xsg1ClJSYy0oZ0XYIxQzZfOqCEwolUeob_AU/edit?usp=sharing')
wtime<-patience.data$sec
hist(wtime,freq=FALSE)
lines(density(wtime),col="red",lwd=2)
#compare it with a theoretical normal distribution curve
curve(dnorm(x,mean=mean(wtime),sd=sd(wtime)),
add=TRUE, col="blue", lwd=2)
legend("topright",col=c("blue","red"),legend =c("estimated normal density curve","kernel density curve"),lwd=2, bty = "n")

Let's fit data to an exponential distribution to the data and check it graphically
require(fitdistrplus)
fit.exp <- fitdist(wtime, "exp")
plot(fit.exp)
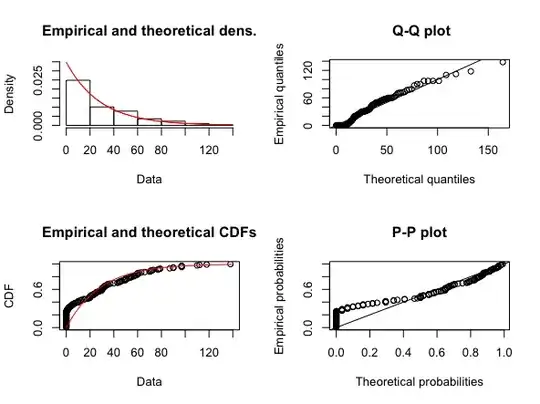
The second and third graph look convincing
Let's fit now the histogram, density curve and exponential curve together
fit.exp#get the estimated rate: 0.03482814
hist(wtime,probability = TRUE)
lines(density(wtime),col="red",lwd=2)
curve(dexp(x, rate = 0.034828136), col = 3, lty = 2,lwd=2,
add=TRUE)
legend("topright",col=c("green","red"),
legend =c("estimated exponential density",
"kernel density"),
lwd=2, bty = "n")