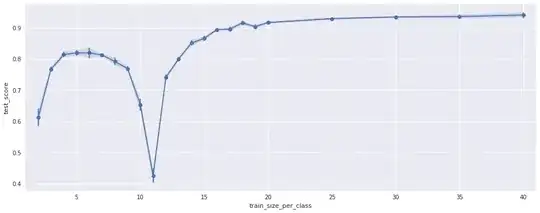
Short story:
I have a classification pipeline consisting of some feature extractors and an LDA classifier. When evaluating the pipeline in a cross-validation I get a decent test accuracy of 94% (for 19 classes). However when evaluating the test accuracy for different amounts of training data I get weird results: the test accuracy overall increases with more training data (which is expected), but for a certain amount of training data the accuracy collapses (see plot):
The plot shows the test accuracy for the pipeline (y-axis) versus the number of samples per class used for training (x-axis). For each of the 19 classes there are 50 training samples. Testing was done on all samples that were not used for training.
The negative peak at 10-11 makes absolutely no sense to me. Can anyone explain this?
The long story:
To make sure this is not a random effect I ran the test 40 times for different, random permutations of the training samples. This is the variance shown in the plot. In addition I repeated the test using only subsets of the features. The results look the same, just the position of the negative peak is different, see the following plots:
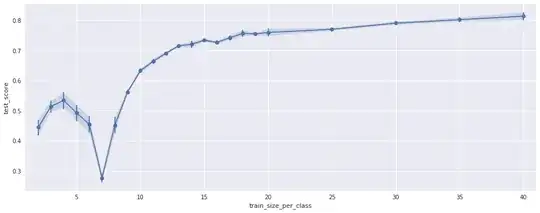 (Using only 110 of the 176 features)
(Using only 110 of the 176 features)
 (Using only the other 66 features)
(Using only the other 66 features)
I noticed that the position of the peak changes with the amout of features, so I ran the test again for 11 different amounts of features. This results in the following plot:
 (x-axis is the same as above. y-axis is the size/16 of the selected featureset, so e.g. 5.0 means 80 features The color is the test accuracy)
(x-axis is the same as above. y-axis is the size/16 of the selected featureset, so e.g. 5.0 means 80 features The color is the test accuracy)
This suggests that there is a direct relation between the position of the peak and the number of features.
Some further points:
- I have quite a lot features (176) for only 19 classes. For sure this is not optimal, but it my opinion it does not explain that peak.
- Some of the variables are collinear (actually a lot of them are). Again, not perfect but does not explain the peak.
Edit:
Here is a plot of the training accuracy:
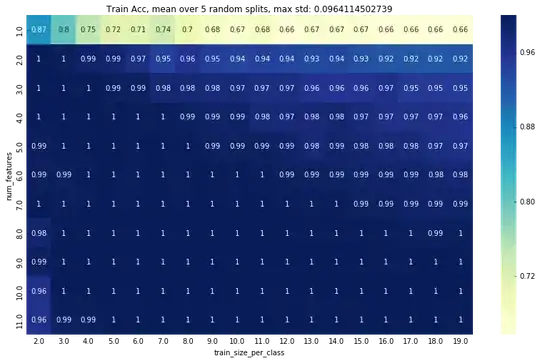
Edit2:
Here is some data. Its calculated using the following settings:
- K = 19 (number of classes)
- p = 192 (number of features)
- n = 2 to 19 (number of training samples PER CLASS, train/test split is a stratified random split)
The outputs are:
- n: number of samples per class
- test data: shape of the test data fed into lda (num samples x num features)
- train data: shape of the train data fed into lda (num samples x num features)
- test and training accuracy
- rank: obtained by printing
rankdefined in https://github.com/scikit-learn/scikit-learn/blob/a95203b/sklearn/lda.py#L369
->
n test data train data test acc train acc rank
2 (912, 192) (38, 192) 0.626 0.947 19
3 (893, 192) (57, 192) 0.775 1.000 38
4 (874, 192) (76, 192) 0.783 1.000 57
5 (855, 192) (95, 192) 0.752 1.000 76
6 (836, 192) (114, 192) 0.811 1.000 95
7 (817, 192) (133, 192) 0.760 1.000 114
8 (798, 192) (152, 192) 0.786 1.000 133
9 (779, 192) (171, 192) 0.730 1.000 152
10 (760, 192) (190, 192) 0.532 1.000 171
11 (741, 192) (209, 192) 0.702 1.000 176
12 (722, 192) (228, 192) 0.727 1.000 176
13 (703, 192) (247, 192) 0.856 1.000 176
14 (684, 192) (266, 192) 0.857 1.000 176
15 (665, 192) (285, 192) 0.887 1.000 176
16 (646, 192) (304, 192) 0.881 1.000 176
17 (627, 192) (323, 192) 0.896 1.000 176
18 (608, 192) (342, 192) 0.913 1.000 176
19 (589, 192) (361, 192) 0.900 1.000 176
20 (570, 192) (380, 192) 0.916 1.000 176
21 (551, 192) (399, 192) 0.907 1.000 176
22 (532, 192) (418, 192) 0.929 0.995 176
23 (513, 192) (437, 192) 0.916 0.995 176
24 (494, 192) (456, 192) 0.909 0.991 176
25 (475, 192) (475, 192) 0.947 0.992 176
26 (456, 192) (494, 192) 0.928 0.992 176
27 (437, 192) (513, 192) 0.927 0.992 176
28 (418, 192) (532, 192) 0.940 0.992 176
29 (399, 192) (551, 192) 0.952 0.991 176
30 (380, 192) (570, 192) 0.934 0.989 176
31 (361, 192) (589, 192) 0.922 0.992 176
32 (342, 192) (608, 192) 0.930 0.990 176
33 (323, 192) (627, 192) 0.929 0.989 176
34 (304, 192) (646, 192) 0.947 0.986 176
35 (285, 192) (665, 192) 0.940 0.986 176
36 (266, 192) (684, 192) 0.940 0.993 176
37 (247, 192) (703, 192) 0.935 0.989 176
38 (228, 192) (722, 192) 0.939 0.985 176
39 (209, 192) (741, 192) 0.923 0.988 176