This question consists of two - but possibly related - parts.
Question #1
We have data on the occurrence of a disease in several age groups over several years. We try to investigate the secular trend of the disease (i.e. how it changes over the years), we will use negative binomial regression with splines for that end, but as the age composition of the (background) population is substantially changing due to aging, we have to apply some adjustment. I can think of two ways:
- Classical method: perform a usual indirect standardization, and add the expected counts as offset to the regression.
- Modern method: don't collapse age groups, feed the whole database to the regression, but add age as a predictor (and the offset is simply to population count of the stratum).
Apart from being ''classical'' and ''modern'', what are the pros and cons of these approaches?
Question #2
Let's try out these two approaches on a simulated dataset.
We first simulate changing age composition (by transiting from Kazakhstan's to Sweden's population pyramid), and add some predictors to create a more or less realistic scenario:
library( data.table )
library( mgcv )
library( epitools )
set.seed( 12 )
RawData <- data.table( read.table( "http://data.princeton.edu/eco572/datasets/preston21long.dat",
header = FALSE, col.names = c( "country", "ageg", "pop", "deaths" ) ) )
RawData$ageg <- rep( c( 0, 1, seq( 5, 85, 5 ) ), 2 )
RawData <- RawData[ , approx( x = c( 0, 20*360-1 ), y = pop,
xout = 0:(20*360-1) ) , .( ageg ) ]
names( RawData )[ c( 2, 3 ) ] <- c( "date", "pop" )
RawData$year <- floor( RawData$date/360 )+1
RawData$month <- floor( ( RawData$date-(RawData$year-1)*360)/30 )+1
RawData$count <- rpois( nrow( RawData ),
exp( log( 0.0001*RawData$pop ) + 0.00004*RawData$ageg^2 -
0.005*(RawData$date/300)^2 + sin( RawData$month/12*2*pi ) ) )
I.e. we have daily data for 20 years.
First let's try the modern approach:
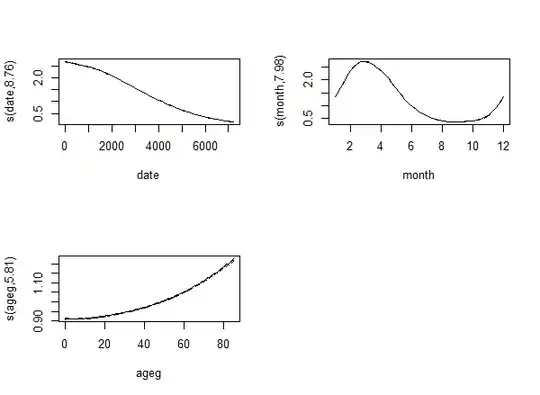
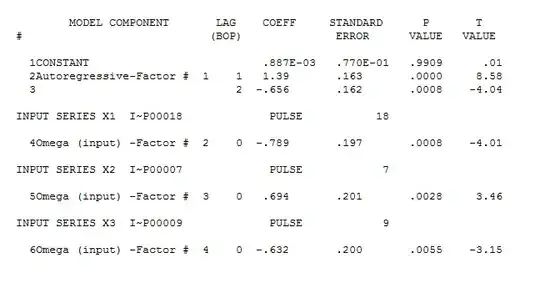
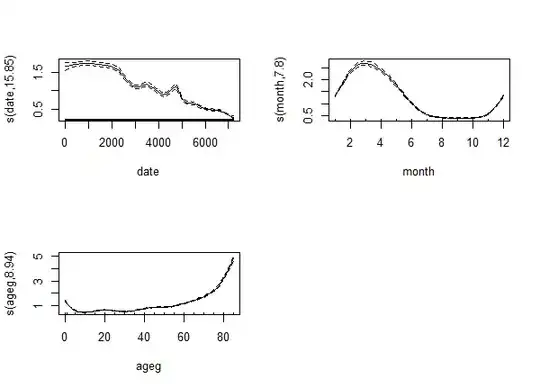
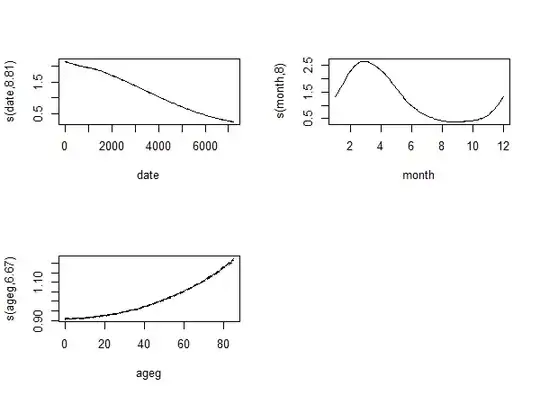
fit <- gam( count ~ s( date ) + s( month, bs = "cc" ) + s( ageg ), offset = log( pop ),
data = RawData, family = poisson( link = "log" ) )
par( mfrow = c( 2, 2 ) )
plot( fit, trans = exp, scale = 0 )
The result is:
Let's now try the classical approach:
StdPop <- RawData[ , .( count = sum( count ), pop = sum( pop ) ), by = .( ageg ) ]
fit2 <- gam( count ~ s( date ) + s( month, bs = "cc" ), offset = log( exp ),
data = merge( RawData, StdPop, by = "ageg" )[
, .( exp = ageadjust.indirect( count.x, pop.x, count.y, pop.y )$sir[ c( "exp" ) ],
count = sum( count.x ) ),
.( date, year, month ) ], family = poisson( link = log ) )
par( mfrow = c( 1, 2 ) )
plot( fit2, trans = exp, scale = 0 )
Result:
As expected, we couldn't obtain information on the age's effect, but otherwise the results are pretty similar. But here comes the surprising part.
Let's say we are worried whether the spline for the time trend was flexible enough (i.e. k is correctly chosen). The standard approach to this problem in mgcv is simply to set k high enough - if k is too low, we have a problem, but if it is high enough, it's exact value usually doesn't really matter.
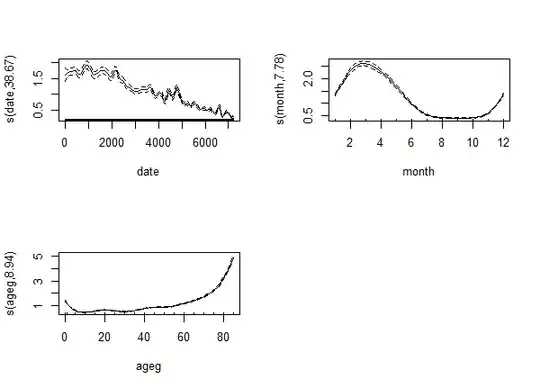
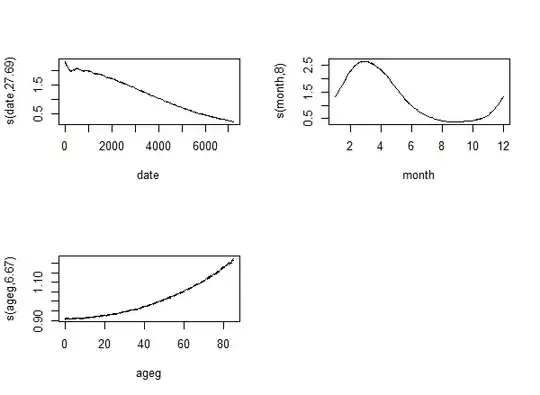
So let's increase k to 30. With the modern approach we see that the effective df increases:
Just as with the traditional approach:
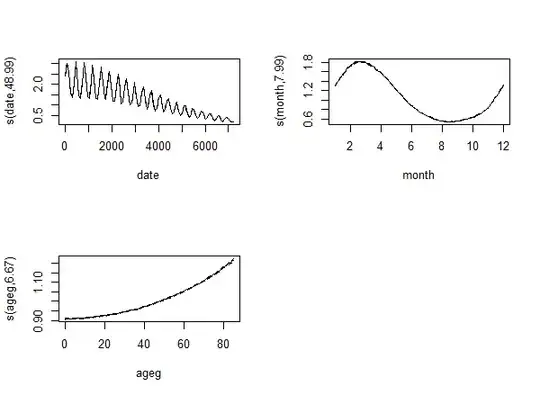
Was 30 still too low? Let's increase it to 50! The modern one produces an even stranger output:
Just as the traditional one:
Something clearly went very wrong with k=50, but even if we put that aside, it is still strange: the effective df - instead of settling when the k is high enough - simply keeps increasing...
What's going on here...?
EDIT (01-Feb-2018)
See earlier attempts here (chronologically).
In the light of an earlier answer, I tried to experiment with the adaptive spline (with bs="ad"), but it didn't really help. For k=30:
With k=50:
EDIT (15-Dec-2017)
Following the suggestions of @Paul, I tried to experiment with including population as a true offset:
RawData$count2 <- round( exp( log( 0.0001*RawData$pop ) + 0.00004*RawData$ageg^2 -
0.005*RawData$year^2 + sin( RawData$month/12*2*pi ) + rnorm( nrow( RawData ), 0, 2 ) ) )
However, it doesn't seem to change the story:
fit <- gam( count2 ~ s( date, k = 30 ) + s( month, bs = "cc" ) + s( ageg ),
offset = log( pop ), data = RawData, family = nb( link = "log" ) )
par( mfrow = c( 2, 2 ) )
plot( fit, trans = exp, scale = 0 )
results in
With k=50:
So it seems that everything is unchanged...
EDIT (08-Jan-2018)
Following another suggestion of @Paul, I tried to change the method, but to no avail...
REML (with the usual k=30):
ML (with the usual k=30):
EDIT (10-Jan-2018)
Following the final suggestion from @Paul I tried to generate the simulated data so that it follows Poisson distribution and set the model also to Poisson:
RawData$count3 <- rpois( nrow( RawData ), exp( log( 0.0001*RawData$pop ) +
0.00004*RawData$ageg^2 - 0.005*RawData$year^2 + sin( RawData$month/12*2*pi ) ) )
fit <- gam( count3 ~ s( date, k = 50 ) + s( month, bs = "cc" ) + s( ageg ),
offset = log( pop ), data = RawData, family = poisson( link = "log" ), method = "ML" )
...but it didn't help either.
k=10:
k=30:
k=50:
So, as before, edf seems to follow k, instead of settling when k is sufficiently high.