Is it possible to specify a variance matrix for the random effects in PROC MIXED with the only restriction that the diagonal entries are equal ? I took a look in the help and I have not found, but this seems strange because such a variance structure is rather natural in some applications.
(EDIT) Consider for instance such a dataset:
> dat
Subject Dose y
1 1 A 10
2 1 A 11
3 1 A 12
4 1 B 30
5 1 B 31
6 1 B 32
7 1 C 100
8 1 C 101
9 1 C 102
10 2 A 11
11 2 A 14
12 2 A 13
13 2 B 33
14 2 B 37
15 2 B 36
16 2 C 105
17 2 C 110
18 2 C 109
19 3 A 9
20 3 A 11
21 3 A 12
22 3 B 30
23 3 B 35
24 3 B 32
25 3 C 115
26 3 C 101
27 3 C 102
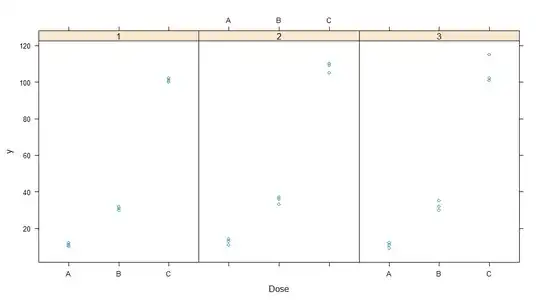
and the following model:
PROC MIXED DATA=dat ;
CLASS SUBJECT DOSE ;
MODEL y = DOSE ;
RANDOM DOSE / subject=SUBJECT type=MYMATRIX ;
RUN; QUIT;
I want a matrix "MYMATRIX" with the same variance for each level of the DOSE factor, but not a compound symmetry matrix because the correlation between the means of the levels are different.
(EDIT2) The mathematical meaning of this model is the following one. Denoting by $i$ the index for the dose level and by $j$ the index for the subject, one has $$(y_{ijk} | \mu_{ij}) \sim_{\text{iid}} {\cal N}(\mu_{ij}, \sigma^2_w), \quad k=1, \ldots, 3 \quad \text{ for all } i,j$$ and $$ \begin{pmatrix} \mu_{1j} \\ \mu_{2j} \\ \mu_{3j} \end{pmatrix} \sim_{\text{iid}} {\cal N}_3 \left( \begin{pmatrix} \mu_1 \\ \mu_2 \\ \mu_3 \end{pmatrix}, G \right), \quad j=1, \ldots, 3$$ The diagonal entries of the $G$ matrix are the between variances for each level of the dose. I want $G$ to be of the form $$G=\begin{pmatrix} \sigma^2_b & \sigma_{12} & \sigma_{13} \\ \sigma_{12} & \sigma^2_b & \sigma_{23} \\ \sigma_{13} & \sigma_{23} & \sigma^2_b \end{pmatrix}$$