Following the links in parenthesis (link1, link2) I wrote a bit of code in MATLAB to simulate a PCA.
After running the code and plotting the results I obtain this graph:
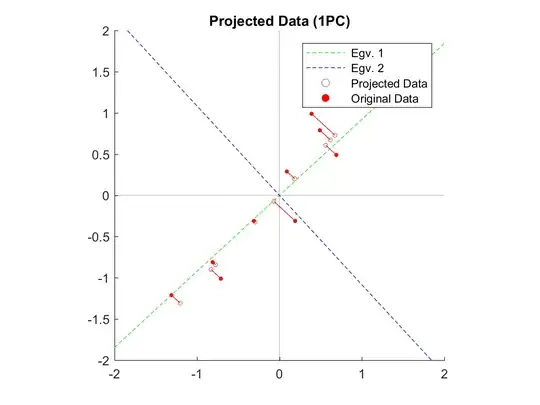
My question is:
The numerical computation seems right. What is the problem with my plot?
Edit1: Below is the full code I used. Please help me finding the bug if you can.
Edit2: Show code output at each step.
Edit3: The slopes of the eigenvectors are [0.9221 -1.0845] while the slopes of the projection lines are [-1.0845]. So they are orthogonal (-1.0845 = -1/0.9221). Below is the bit of code to get the two principal components axes. Where am I wrong?
Edit4: Solved; I was calculating the inverse of m (eigen slopes). Changing m = eigenvectors(1, :) ./ eigenvectors(2,:); to m = eigenvectors(2, :) ./ eigenvectors(1,:); solve the problem and return the right graph. Thanks to amoeba for pointing that out.
m = eigenvectors(1, :) ./ eigenvectors(2,:);
xe = linspace(-2, 2, 3);
ye1 = m(1) .* xe;
ye2 = m(2) .* xe;
E1 = plot(xe, ye1, '-- g', 'DisplayName',' Egv. 1');
E2 = plot(xe, ye2, '-- b', 'DisplayName',' Egv. 2');
Data
x = [2.5 0.5 2.2 1.9 3.1 2.3 2 1 1.5 1.1];
y = [2.4 0.7 2.9 2.2 3.0 2.7 1.6 1.1 1.6 0.9];
xm = x - mean(x);
ym = y - mean(y);
[xm; ym]'
0.690000000000000 0.490000000000000
-1.310000000000000 -1.210000000000000
0.390000000000000 0.990000000000000
0.090000000000000 0.290000000000000
1.290000000000000 1.090000000000000
0.490000000000000 0.790000000000000
0.190000000000000 -0.310000000000000
-0.810000000000000 -0.810000000000000
-0.310000000000000 -0.310000000000000
-0.710000000000000 -1.010000000000000
Covariance matrix
cov_mat = cov(x, y);
cov_mat =
0.616555555555556 0.615444444444444
0.615444444444444 0.716555555555556
Get Eigenvalues and Eigenvectors and sort
[eigenvectors , eigenvalues] = eig(cov_mat, 'vector');
[~, I] = sort(eigenvalues, 1, 'descend');
eigenvalues = eigenvalues(I,:);
eigenvectors = eigenvectors(:,I);
eigenvalues =
1.284027712172784
0.049083398938327
eigenvectors =
0.677873398528012 -0.735178655544408
0.735178655544408 0.677873398528012
Transform
FinalData1 = [xm; ym]' * eigenvectors(:,1); % one PC
FinalData1 =
0.827970186201088
-1.777580325280429
0.992197494414889
0.274210415975400
1.675801418644540
0.912949103158808
-0.099109437498444
-1.144572163798660
-0.438046136762450
-1.223820555054740
Re-project onto the original dimensions
Projections = FinalData1 * eigenvectors(:,1)';
Projections =
0.561258964000002 0.608706008322169
-1.204974416254373 -1.306839113661857
0.672584287549999 0.729442419978468
0.185879946589024 0.201593644953067
1.135981202914638 1.232013433918505
0.618863911241362 0.671180694240766
-0.067183651223270 -0.072863143011869
-0.775875022534758 -0.841465024555053
-0.296939823439228 -0.322042169891440
-0.829595398843395 -0.899726750292755
Plot
m = eigenvectors(1, :) ./ eigenvectors(2,:);
xe = linspace(-2, 2, 3);
ye1 = m(1) .* xe;
ye2 = m(2) .* xe;
data = [xm; ym]';
figure, s1 = scatter(Projections(:, 1), Projections(: ,2), 10, 'r', 'DisplayName',' Projected Data'); title('Projected Data (1PC)');
hold on
xlim([-2 2]);
ylim([-2 2]);
E1 = plot(xe, ye1, '-- g', 'DisplayName',' Egv. 1');
E2 = plot(xe, ye2, '-- b', 'DisplayName',' Egv. 2');
plot(0.*xe, xe, ': k');
plot(xe, 0.*xe, ': k');
s2 = scatter(xm, ym, 10, 'filled', 'r', 'DisplayName',' Original Data');
slopes = zeros(size(data,1),1);
pj = [Projections(:, 2), Projections(: ,1)];
for i = 1:size(data,1)
l = [data(i,:); pj(i,:)];
slopes(i) = (l(2,1) - l(1,1)) / (l(2,2) - l(1,2));
plot(l(:,1), l(:,2), 'r')
end
legend([E1 E2 s1 s2])
pbaspect([1 1 1])
hold off
