You can look at wikipedia.
The hypergeometric test uses the hypergeometric distribution to measure the statistical significance of having drawn a sample consisting of a specific number of k successes (out of n total draws) from a population of size N containing K successes. In a test for over-representation of successes in the sample, the hypergeometric p-value is calculated as the probability of randomly drawing k or more successes from the population in n total draws. In a test for under-representation, the p-value is the probability of randomly drawing k or fewer successes.
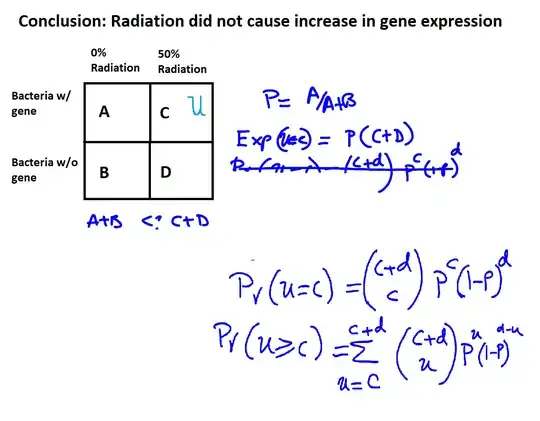
The null-hypothesis here is that u, that, is, the probability under radation of the gene, is equal to p, the probability without radiation of the gene. While I would think that p has to be considered uncertain here as well, here a shortcut is taken by estimating p directly from the sample, as $\frac{A}{A+B}$. It is possible that this is equivalent.
The formulas are the probability under the null hypothesis (where $p = \frac{A}{A + B}$) that you would see $C$ or more bacteria with the gene out of $C + D$ samples. So that is the $p$-value.
That is the answer. Since you are thinking about $p$-values, maybe you can read a couple of blogposts from Andrew Gelman, I think it is a good idea to be a bit sceptical about this hypothesis framework.