Hello to the CrossValidated community. I could use some help.
I am struggling with the analysis of an experiment, specifically with deciding and defining in code the structure of a mixed model for analysing it.
The experimental design is as follows:
We wanted to see if there are any differences on the growth of lesions caused by different combinations of pathogens (Treatment in the data).
To investigate this we used detached leaves and inoculated them with said Treatments. Each leaf was inoculated in two spots which were averaged to give us the Area of each lesion. Measurements were done per day for all leaves in the experiment, for days 4-9 after the inoculation (Day in the data below).
Two difference cultivars of potato plants were used in the experiment (Cultivar in the data).
Leaves were arranged in boxes (Box in the data) for a complete randomized block design. Each box contained 7 leaves of each cultivar, each leaf inoculated with one of the Treatment, so a total of 14 leaves in each box.
LeafID is a unique identifier for each of leaves in the experiment.
My understanding is that Box, Cultivar, Treatment and Day are crossed factors. LeafID is crossed with Day and Box and nested within Treatment and Cultivar. I am mentioning this to check my understanding of the experimental design and furthermore to understand if it's important for defining the model. I seem to remember reading that lmer accounts for nested or crossed designs internally but can't quote a source unfortunately.
Here is a pick at my data structure in R, Day and Area are coded as double, all other variables are coded as factors
Box Cultivar Treatment Day LeafID Area
1 Arsenal Control 4 121 0.0000000
1 Arsenal Control 5 121 0.0000000
1 Arsenal Control 6 121 0.0000000
1 Arsenal Control 7 121 0.0000000
1 Arsenal Control 8 121 0.0000000
1 Arsenal Control 9 121 0.0000000
1 Arsenal A.solani 4 21 0.1745344
1 Arsenal A.solani 5 21 0.3062256
1 Arsenal A.solani 6 21 0.3721086
1 Arsenal A.solani 7 21 0.4217482
....
My understanding is that this can be analysed as either a repeated measures or a time-series design, and the choice really depends on the research question. I chose to look at this as a repeated measures design, mostly because it's easier for me to grasp the concepts and statistics involved. I am more than happy to hear differing opinions of course!
I tried to fit my model the same way Crawley does in the R Book (p,695, "Mixed-effects models with temporal pseudoreplication")
Initially we came up with the following model:
m0.full <- lme(data=DLA2, Area ~ Treatment * Cultivar * Day ,
random = ~Day | LeafID, na.action = na.exclude ,method = "ML")
The difference from what Crawley does is that our model has extra fixed effects and their interactions Treatment * Cultivar * Day and we are using Day and its interaction as we care about the progress over time and how this affects our growth.
What I am struggling with is deciding if Cultivar should be a fixed or random effect and if including Day in the fixed effects also gives the wrong interpretation.
I've spent countless hours going through mixed model tutorials and I know that going for a very complex model is not the way to go. I guess the most confusing part is deciding what to have as fixed and random. I've seen some people suggesting you can use a factor in both fixed and random parts of the model but i have trouble grasping the concept.
I would be happy to share the entire dataset if that helps, any suggestions of the best way to do this would be welcome.
I am looking forward for some suggestions on how to handle my analysis and what could be improved.
I would be happy using either lme or lmer.
Thank you all for your time!
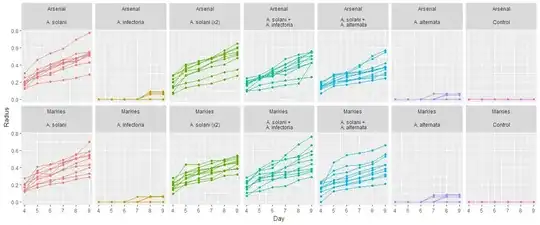
EDIT: I added a plot of my data for each different Treatment*Cultivar combination. Each panel shows the full replicates corresponding to the combination
UPDATE
I came up with these two models, mod.rep takes into account the repeated measures of the Day variable, while mod.time uses a time-series approach as used by Crawley(The R Book). I also accounted for heteroscedasticity using the varIdent function.
Are those models correct ?
mod.rep <- lme(data=DLA2, Radius ~ Treatment * Cultivar, random = ~
Day|LeafID, na.action = na.omit ,method = "ML", weights =
varIdent(form=~1|Treatment))
mod.time <- lme(data=DLA2, Radius ~ Day * Treatment * Cultivar, random =
~1|LeafID, na.action = na.exclude ,method = "ML", weights =
varIdent(form=~1|Treatment))
I still have some trouble translating the random effects formulas into words...