The t-statistic can have next to nothing to say about the predictive ability of a feature, and they should not be used to screen predictor out of, or allow predictors into a predictive model.
P-values say spurious features are important
Consider the following scenario setup in R. Let's create two vectors, the first is simply $5000$ random coin flips:
set.seed(154)
N <- 5000
y <- rnorm(N)
The second vector is $5000$ observations, each randomly assigned to one of $500$ equally sized random classes:
N.classes <- 500
rand.class <- factor(cut(1:N, N.classes))
Now we fit a linear model to predict y given rand.classes.
M <- lm(y ~ rand.class - 1) #(*)
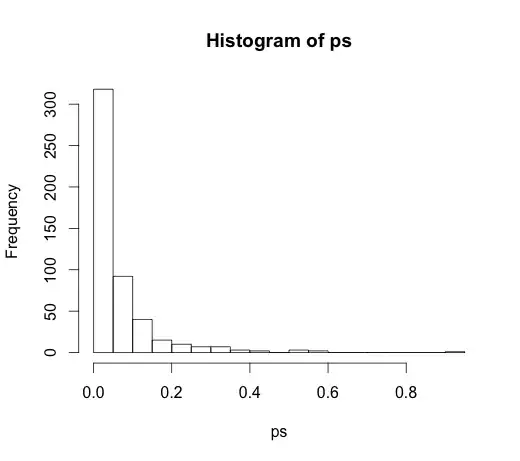
The correct value for all of the coefficients is zero, none of them have any predictive power. None-the-less, many of them are significant at the 5% level
ps <- coef(summary(M))[, "Pr(>|t|)"]
hist(ps, breaks=30)
In fact, we should expect about 5% of them to be significant, even though they have no predictive power!
P-values fail to detect important features
Here's an example in the other direction.
set.seed(154)
N <- 100
x1 <- runif(N)
x2 <- x1 + rnorm(N, sd = 0.05)
y <- x1 + x2 + rnorm(N)
M <- lm(y ~ x1 + x2)
summary(M)
I've created two correlated predictors, each with predictive power.
M <- lm(y ~ x1 + x2)
summary(M)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.1271 0.2092 0.608 0.545
x1 0.8369 2.0954 0.399 0.690
x2 0.9216 2.0097 0.459 0.648
The p-values fail to detect the predictive power of both variables because the correlation affects how precisely the model can estimate the two individual coefficients from the data.
Inferential statistics are not there to tell about the predictive power or importance of a variable. It is an abuse of these measurements to use them that way. There are much better options available for variable selection in predictive linear models, consider using glmnet.
(*) Note that I am leaving off an intercept here, so all the comparisons are to the baseline of zero, not to the group mean of the first class. This was @whuber's suggestion.
Since it led to a very interesting discussion in the comments, the original code was
rand.class <- factor(sample(1:N.classes, N, replace=TRUE))
and
M <- lm(y ~ rand.class)
which led to the following histogram