for example the data has a binary response y, a numerical predictor week, and some categorical/factor predictors: ap, hilo, ID, trt:
y ap hilo week ID trt
1 y p hi 0 X01 placebo
2 y p hi 2 X01 placebo
3 y p hi 4 X01 placebo
4 y p hi 11 X01 placebo
5 y a hi 0 X02 drug+
6 y a hi 2 X02 drug+
7 n a hi 6 X02 drug+
8 y a hi 11 X02 drug+
9 y a lo 0 X03 drug
10 y a lo 2 X03 drug
Logistic regression result looks like:
Coefficients: (4 not defined because of singularities)
Estimate Std. Error z value Pr(>|z|)
(Intercept) 2.550e+00 1.251e+00 2.038 0.041561 *
app 1.924e+01 8.359e+03 0.002 0.998164
hilolo -1.562e+00 1.617e+00 -0.966 0.334074
week -2.127e-01 6.377e-02 -3.335 0.000852 ***
IDX02 -2.525e-01 1.721e+00 -0.147 0.883337
IDX03 2.081e+01 7.562e+03 0.003 0.997804
IDX04 1.572e+00 1.127e+04 0.000 0.999889
IDX05 1.572e+00 1.127e+04 0.000 0.999889
...
Null deviance: 217.38 on 219 degrees of freedom
Residual deviance: 118.51 on 169 degrees of freedom
AIC: 220.51
Number of Fisher Scoring iterations: 19
It tells us for predictor ID, the baseline is ID == 'X01' (not shown in result). Comparing ID == 'X02' to '01', the change is not significant, because the p-value is 0.883337.
My question is how do you compare ID == 'X02' to 'X03'? The log adds changes by 2.081e+01 - (-2.525e-01), what error do I compare this difference to, and how to calculate p value?
Can somebody give an example using ID == 'X02' to 'X03' please? Thank you.
I leave the code later because my question is only about theory. Code in R:
library(MASS)
library(stats)
data('bacteria')
dat = bacteria
glm_model = glm(y ~ ., family = binomial, data = dat)
summary(glm_model)
What I know (not very sure, and would like to get verified)
each coefficient has it's estimate and std.error, because there's a population of many different values of this coefficient calculated by using differently sampled data.
So comparing ID == 'X02' to 'X03' is to compare the mean of two populations. I read this post, so 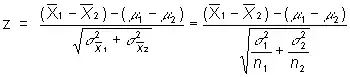
so, is delta(x1bar) in the equation the std.error in the glm result? I don't need to divide by n any more, right (since the std.error in the glm result in already the standard deviation of the mean)?
Comparing ID == 'X02' to 'X03':
z = (-2.525e-01 - 2.081e+01) / sqrt(1.721e+00^2 + 7.562e+03^2)
= -0.002785
is this correct? Thanks-