Intuitively, there's no problem because for each $k=1, 2, \ldots, n$ we may pair the variable of mean $k/n$ with that of mean $(n-k)/n$ (except possibly when $2k=n,$ which will not matter asymptotically). The sum of these variables has mean $1/2$ and (obviously) has no skewness. The variances of these sums are
$$\frac{k}{n}\left(1 - \frac{k}{n}\right) + \left(1 - \frac{k}{n}\right)\frac{k}{n} = 2\frac{k}{n}\left(1 - \frac{k}{n}\right)$$
which range from $2/n^2$ to $1/2.$ This range might give us pause, but asymptotically it causes no problems: when we standardize the sum (its mean is $\mu(n)=(1+n)/2$ and its variance is $\sigma^2(n)=(n^2-1)/(6n),$ both elementary computations) it converges fairly rapidly to a standard Normal distribution. The excess kurtosis is always negative (the tails are obviously limited in extent) but shrinks to zero.
A rigorous demonstration computes that the cumulant generating function of the standardized sum is $t^2/2 + O(n^{-1})O(t^4)$ which converges as $n$ grows large to the c.g.f. of the standard Normal distribution.
As a check, this R simulation generates 10,000 such sums for $n=20$ and visually compares their distribution to the Normal distribution with parameters $\mu(20),\sigma^2(20):$
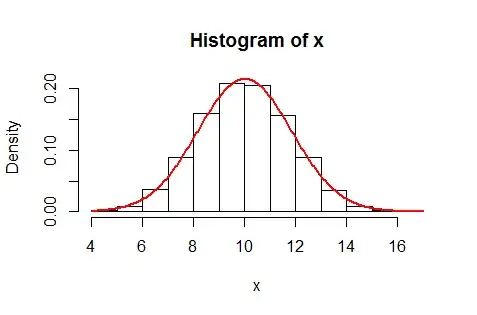
The red curve plots the corresponding theoretical probabilities. It is (already) an excellent fit.
n <- 20
n.sim <- 1e4
set.seed(17)
p <- (1:n)/n
x <- colSums(matrix(rbinom(n*n.sim, 1, p), nrow=n))
hist(x, freq=FALSE, breaks=min(x):max(x))
m <- sum(p)
v <- sum(p * (1-p))
curve(pnorm(x+1, m, sqrt(v)) - pnorm(x, m, sqrt(v)), col="Red", add=TRUE, lwd=2, n=301)
