Edited after helpful feedback from Mark White (thank you!)
One option is to calculate both relationships (B1 with A, and B2 with A) in a single model that also estimates the difference between them. This is easy to accomplish with multiple regression. You would run a model with A as the dependent variable, and then one continuous variable with all of the scores for B1 and B2, a categorical variable indicating which variable it was (B1 or B2), and the interaction between them. In r:
> set.seed(24601)
>
> library(tidyverse)
> library(mvtnorm)
> cov <- matrix(c(1, .4, .16,.4, 1, .4, .16, .4, 1), ncol=3, byrow=TRUE)
> mydata <- rmvnorm(n=100, sigma = cov)
> colnames(mydata) = c("A", "B1", "B2")
> head(mydata)
A B1 B2
[1,] -0.1046382 0.6031253 0.5641158
[2,] -1.9303293 -0.7663828 -0.7921836
[3,] 0.1244192 -0.4413581 -1.2376256
[4,] -3.2822601 -1.2512055 -0.5586773
[5,] -0.9543368 -0.1743740 1.1884185
[6,] -0.4843183 -0.2612668 -0.7161938
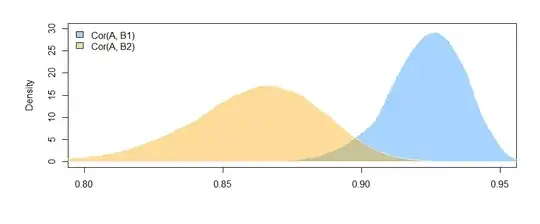
Here are the correlations from the data I generated:
> cor(mydata)
A B1 B2
A 1.0000000 0.4726093 0.3043496
B1 0.4726093 1.0000000 0.3779376
B2 0.3043496 0.3779376 1.0000000
>
Changing the format of the data to meet the needs of the model (reformatting to "long"):
> mydata <- as.data.frame(mydata) %>%
+ gather("var", "value", B1, B2)
>
Here's the model:
summary(lm(A~value*var, data = mydata))
Call:
lm(formula = A ~ value * var, data = mydata)
Residuals:
Min 1Q Median 3Q Max
-2.89310 -0.52638 0.02998 0.64424 2.85747
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.09699 0.09014 -1.076 0.283
value 0.47445 0.09305 5.099 8.03e-07 ***
varB2 -0.10117 0.12711 -0.796 0.427
value:varB2 -0.13256 0.13965 -0.949 0.344
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.891 on 196 degrees of freedom
Multiple R-squared: 0.158, Adjusted R-squared: 0.1451
F-statistic: 12.26 on 3 and 196 DF, p-value: 2.194e-07
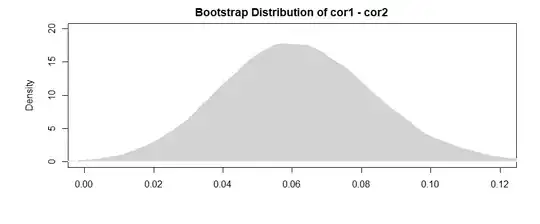
The results here (from my made-up data) suggest that there is a significant relationship between B1 and A (the test of the "value" coefficient, since B1 is the reference group for the "var" coefficient), but that the difference between the B1 relationship with A and the B2 relationship with A is not significant (the test of the "value:varB2" coefficient).
If you like thinking in terms of correlation rather than regression coefficients, just standardize all of your variables (A, B1, and B2) before running the model and the regression coefficients you'll get will be standardized (not quite the same thing as a zero-order correlation, but much closer in terms of interpretation).
Also note that this will restrict your analysis to just the cases that have both B1 and B2 (listwise deletion). As long as that leaves you with enough data to not be underpowered, and as long as the missing data are missing randomly (or a small enough proportion of the total data to not matter much even if they are missing nonrandomly), then that's fine.
The fact that you're restricting your analysis to the same dataset for estimating effects for both B1 and B2 (rather than using slightly different datasets, based on the different patterns of missingness) has the advantage of making interpretation of the difference between correlations a little more straightforward. If you calculate the correlations separately for each and then test the difference between them, you run into the problem that the underlying data are slightly different in each case --- any difference you see could be due to differences in the samples as much as differences in the actual relationships between variables.