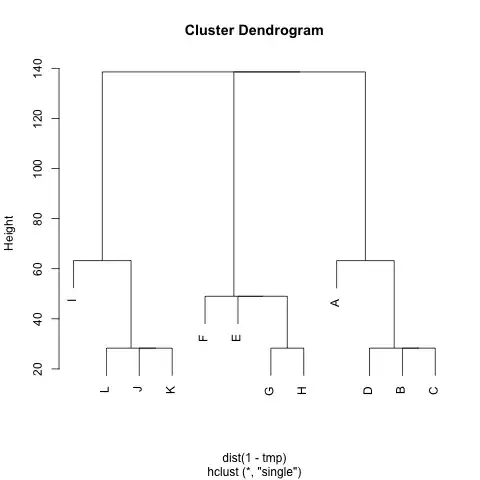
I have a (symmetric) matrix M that represents the distance between each pair of nodes. For example,
A B C D E F G H I J K L
A 0 20 20 20 40 60 60 60 100 120 120 120
B 20 0 20 20 60 80 80 80 120 140 140 140
C 20 20 0 20 60 80 80 80 120 140 140 140
D 20 20 20 0 60 80 80 80 120 140 140 140
E 40 60 60 60 0 20 20 20 60 80 80 80
F 60 80 80 80 20 0 20 20 40 60 60 60
G 60 80 80 80 20 20 0 20 60 80 80 80
H 60 80 80 80 20 20 20 0 60 80 80 80
I 100 120 120 120 60 40 60 60 0 20 20 20
J 120 140 140 140 80 60 80 80 20 0 20 20
K 120 140 140 140 80 60 80 80 20 20 0 20
L 120 140 140 140 80 60 80 80 20 20 20 0
Is there any method to extract clusters from M (if needed, the number of clusters can be fixed), such that each cluster contains nodes with small distances between them. In the example, the clusters would be (A, B, C, D), (E, F, G, H) and (I, J, K, L).
I've already tried UPGMA and k-means but the resulting clusters are very bad.
The distances are the average steps a random walker would take to go from node A to node B (!= A) and go back to node A. It's guaranteed that M^1/2 is a metric. To run k-means, I don't use the centroid. I define the distance between node n cluster c as the average distance between n and all nodes in c.
Thanks a lot :)