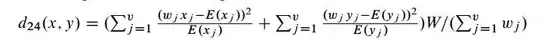I have a couple straightforward questions and hope I can get some straightforward suggestions. I would like to perform a cluster analysis on species counts that are recorded in sequential time blocks so that I can group species into guilds or communities based on temporal co-occurrence (let's assume I have no useful prior information on these species). As you might have already guessed, pairwise euclidean distance is not getting the job done as it does not incorporate temporal similarity. So, I want to compute my own distance matrix to use in hierarchical clustering. Below is an example of a pair of species counts, but there are over 100 species.
What distance or similarity measure would be best suited to this situation? I'm just sort of throwing this one out there. Note there are a lot of 0's in the abundance matrix.
IF I were to use a chi-square distance:
0's end up in the denominator and a blank matrix is returned (this happens regardless of the software I use). I like the idea of the chi-square distance, so how can I get the software to assign a distance of 1 in the situation where 0 is in the denominator, instead of crashing out and returning a blank matrix? I am comfortable using R or SAS to accomplish this, so any little snippet of code to accomplish this simple task would be very helpful.
Any additional thoughts on the actual clustering method would also be helpful.