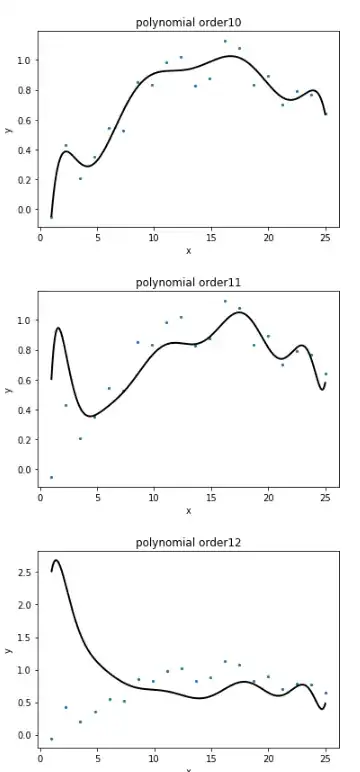
I am fitting a high order polynomial fit (order 15+) to some simulated training data. I know that features become collinear as i increase the order of polynomial but i do not undersand why my fits are so off ! even in case of collinearity the fits should be reasonable. The issue is not related to size of the training data for example see figures below with lots of training samples
The code is below:
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cross_validation import KFold
import matplotlib.pyplot as plt
% matplotlib inline
def gen_data(num_train):
np.random.seed(100)
trainX = np.float64(np.linspace(1,1.5,num_train))
train_noise = np.float64(np.random.normal(0, 0.1, num_train))
trainY = np.sin(10*trainX) + train_noise
return trainX, trainY
def polynomial_feature(X,order):
for i in range(2,order+1):
X = np.column_stack((X,X[:,0]**i))
return X
def append_bias(X):
num_ins, num_feas = X.shape[0], X.shape[1]
Xb = np.ones((num_ins, num_feas+1))
Xb[:, 1:] = X
return Xb
def l2_closed_form(Xb, y):
num_feas = Xb.shape[1]
return np.dot(np.linalg.inv(np.dot(Xb.T, Xb)), np.dot(Xb.T, y))
def plotXY(trainX, trainY, plotX, plotY,title=None):
plt.scatter(trainX[:, 1], trainY, s= 2)
plt.plot(plotX[:, 1], plotY, color = 'k', linewidth=2)
plt.xlabel('x')
plt.ylabel('y')
plt.show()
def q1(trainX, trainY):
theta = l2_closed_form(trainX, trainY)
plotX, _ = gen_data(num_train=1000)
plotX = polynomial_feature(plotX.reshape(-1,1),polynomalOrder)
plotX = append_bias(plotX)
plotY = np.dot(plotX, theta)
plotXY(trainX, trainY, plotX, plotY)
def load_data():
trainX, trainY= gen_data(num_train=15)
trainX = polynomial_feature(trainX.reshape(-1,1),polynomalOrder)
trainX = append_bias(trainX)
return trainX, trainY
polynomalOrder = 15
global polynomalOrder
trainX, trainY = load_data()
q1(trainX, trainY)