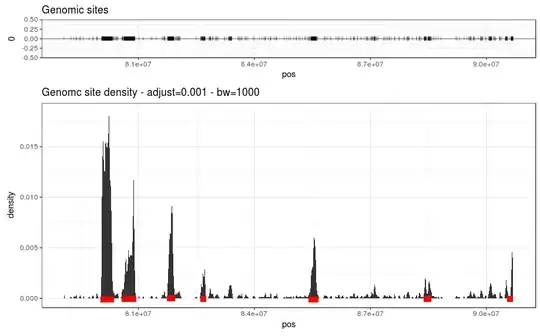
I've ~150,000 genomic position that seems to be clustered in specific genomic regions (hotspot). However these "hotspots" may have different sizes (from very small ~ 10,000bp to very large ~500,000bp - bp = base pair). Could someone give me some advice to detect such peaks ? My idea was to use a small window-based approach and to find adjacent small-windows were the number of positions are significantly higher than random (using simulation).
Here's an subset of my data focuses on a portion of one chromosome. The top panel shows each individual genomic positions of interest (one vertical bar represents one site). The bottom panel shows the density computed using ggplot's stat_density using adjust=0.001 and bw=1000. I manually added the the red lines to show the information I want to extract from such data. An important point would be to extract only peak region that are more dense than by chance. I was thinking to perform a simulation were I randomly distribute 150,000 genomic sites and computes a kind of background density in order to compare with my real data. Any advice ?
Edit : I add the same plot with 5 random set of genomic sites (same size as the real dataset). My idea is to extract these region over the background.
Thanks