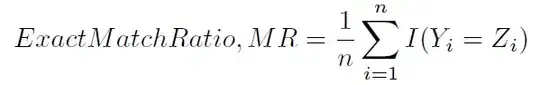The subset accuracy is indeed a harsh metric. To get a sense of how good or bad 0.29 is, some idea:
- look at how many labels you have an average for each sample
- look at the inter-annotator agreement, if available (if not, try yourself to see what subset accuracy the obtained when you are the classifier)
- think whether topic are well defined
- look at how many samples you have for each label
You may also want to compute the hamming score, to see whether your classifier is clueless, or is instead decently good but have issue predicting all labels correctly. See below to compute the hamming score.
At the same time, from what I understand I cannot use scikit.metrics with OneVsRestClassifier so how can I get some metrics (F1,Precision,Recall etc) so as to figure out what is wrong?
See How to compute precision/recall for multiclass-multilabel classification?. I forgot whether sklearn supports it, I recall it had some limitations, e.g. sklearn doesn't support multi-label for confusion matrix. That would be a good idea to see these numbers indeed.
Hamming score:
In a multilabel classification setting, sklearn.metrics.accuracy_score only computes the subset accuracy (3): i.e. the set of labels predicted for a sample must exactly match the corresponding set of labels in y_true.
This way of computing the accuracy is sometime named, perhaps less ambiguously, exact match ratio (1):
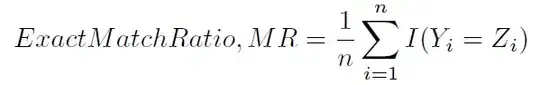
Another typical way to compute the accuracy is defined in (1) and (2), and less ambiguously referred to as the Hamming score (4) (since it is closely related to the Hamming loss), or label-based accuracy). It is computed as follows:
Here is a python method to compute the Hamming score:
# Code by https://stackoverflow.com/users/1953100/william
# Source: https://stackoverflow.com/a/32239764/395857
# License: cc by-sa 3.0 with attribution required
import numpy as np
y_true = np.array([[0,1,0],
[0,1,1],
[1,0,1],
[0,0,1]])
y_pred = np.array([[0,1,1],
[0,1,1],
[0,1,0],
[0,0,0]])
def hamming_score(y_true, y_pred, normalize=True, sample_weight=None):
'''
Compute the Hamming score (a.k.a. label-based accuracy) for the multi-label case
https://stackoverflow.com/q/32239577/395857
'''
acc_list = []
for i in range(y_true.shape[0]):
set_true = set( np.where(y_true[i])[0] )
set_pred = set( np.where(y_pred[i])[0] )
#print('\nset_true: {0}'.format(set_true))
#print('set_pred: {0}'.format(set_pred))
tmp_a = None
if len(set_true) == 0 and len(set_pred) == 0:
tmp_a = 1
else:
tmp_a = len(set_true.intersection(set_pred))/\
float( len(set_true.union(set_pred)) )
#print('tmp_a: {0}'.format(tmp_a))
acc_list.append(tmp_a)
return np.mean(acc_list)
if __name__ == "__main__":
print('Hamming score: {0}'.format(hamming_score(y_true, y_pred))) # 0.375 (= (0.5+1+0+0)/4)
# For comparison sake:
import sklearn.metrics
# Subset accuracy
# 0.25 (= 0+1+0+0 / 4) --> 1 if the prediction for one sample fully matches the gold. 0 otherwise.
print('Subset accuracy: {0}'.format(sklearn.metrics.accuracy_score(y_true, y_pred, normalize=True, sample_weight=None)))
# Hamming loss (smaller is better)
# $$ \text{HammingLoss}(x_i, y_i) = \frac{1}{|D|} \sum_{i=1}^{|D|} \frac{xor(x_i, y_i)}{|L|}, $$
# where
# - \\(|D|\\) is the number of samples
# - \\(|L|\\) is the number of labels
# - \\(y_i\\) is the ground truth
# - \\(x_i\\) is the prediction.
# 0.416666666667 (= (1+0+3+1) / (3*4) )
print('Hamming loss: {0}'.format(sklearn.metrics.hamming_loss(y_true, y_pred)))
Outputs:
Hamming score: 0.375
Subset accuracy: 0.25
Hamming loss: 0.416666666667
(1) Sorower, Mohammad S. "A literature survey on algorithms for multi-label learning." Oregon State University, Corvallis (2010).
(2) Tsoumakas, Grigorios, and Ioannis Katakis. "Multi-label classification: An overview." Dept. of Informatics, Aristotle University of Thessaloniki, Greece (2006).
(3) Ghamrawi, Nadia, and Andrew McCallum. "Collective multi-label classification." Proceedings of the 14th ACM international conference on Information and knowledge management. ACM, 2005.
(4) Godbole, Shantanu, and Sunita Sarawagi. "Discriminative methods for multi-labeled classification." Advances in Knowledge Discovery and Data Mining. Springer Berlin Heidelberg, 2004. 22-30.