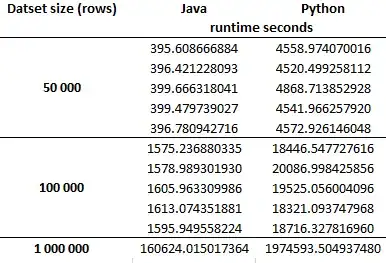
You can think of your data as blocked, or being a two-factor experiment where one factor is a nuisance. These are repeated measures, but they are not paired because the blocks aren't of size $2$. Taking, for example, the data that come from the java code run on a given dataset of 50,000, you have $5$ repeated measures. These are matched to another $5$ repeated measures from the Python code, in the sense that the codes were run on the same dataset. A matched pair would be $1$ datum that corresponds to $1$ datum. You have $5$ data that correspond to $5$ data. Two matched sets of size $5$ each aren't a 'pair'.
In theory, you could use a random effect to account for the non-independence of the data. However, with just three datasets, this is pretty sketchy. In your case, I would control for the non-independence with a fixed factor. In other words, you have two factors in your experiment, one of which (dataset) you don't really care about and only want to control for to account for the non-independence that would otherwise occur.
The question of whether you should make parametric assumptions (like normality) is well addressed in this great CV thread: How to choose between t-test or non-parametric test e.g. Wilcoxon in small samples. If you wanted to use a nonparametric approach, you could use an ordinal logistic regression (cf., What is the non-parametric equivalent of a two-way ANOVA that can include interactions?). If you wanted the parametric equivalent, you could run a standard two-way ANOVA. The ordinal model is probably safer, but it won't matter with your data either way, since the effect is so large.
On a different note, let me address some of the misunderstandings in the question. There are two tests that are sometimes called 'the Wilcoxon test': the Wilcoxon rank sum test, and the Wilcoxon signed rank test. The former is for two independent samples, and is also called the Mann-Whitney U-test. The latter is for two dependent samples. Neither requires that your data are normally distributed. If your data were paired (e.g., if you ran each dataset only once with each software), then you could use the Wilcoxon signed rank test. The complication here is that you don't have paired data.