This is because PCA scores are simply original data in a rotated coordinate frame.
Below on the left I show some example 2D data (100 points in 2D) and on the right the corresponding PCA scores. The data cloud simply gets rotated clockwise by approximately 45 degrees.
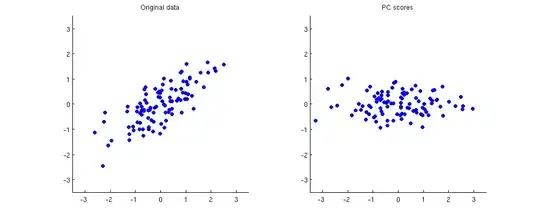
If it is not completely clear to you how one gets from the first subplot to the second one or why PCA amounts to rotation, take a look at our very informative thread Making sense of principal component analysis, eigenvectors & eigenvalues. In my answer there I am using exactly the same toy dataset as displayed here. Some other answers are very much worth reading too.
Now, to your question.
Clustering methods are usually based on Euclidean distances between points. The points that lie close to each other get clustered together; the ones that are far away get assigned to different clusters. As you can see above, all distances between all points stay exactly the same after PCA.
Hence the identical clustering results. Here are both representations clustered with k-means with $k=3$:

As you see, the clustering results are identical.
Can PCA make any difference at all?
Yes. One can use it in two ways:
- Standardize all scores to unit variance; or
- Use only a subset of principal components, usually the ones that explain the most variance.
Here is how it looks like in the same toy example. On the left I am using standardized scores (note how different the clusters become), on the right I am using only PC1.