I am running a multivariate ols model where my dependent variable is Food Consumption Score, an index created by the weighted sum of the consumption occurrences of some given food categories.
Although I have tried different specifications of the model, scaled and/or log-transformed the predictors, the Breusch-Pagan test always detects strong heteroskedasticity.
- I exclude the usual cause of omitted variables;
- No presence of outliers, especially after log-scaling and normalizing;
- I use 3/4 indexes created by applying Polychoric PCA, however even excluding some or all of them from the OLS does not change the Breusch-Pagan output.
- Only few (usual) dummy variables are used in the model: sex, marital status;
- I detect high degree of variation occurring between regions of my sample, despite controlling with the inclusion of dummies for each region and gaining more 20% in terms of adj-R^2, the heteroskedasticity reamins.
- The sample has 20,000 observations.
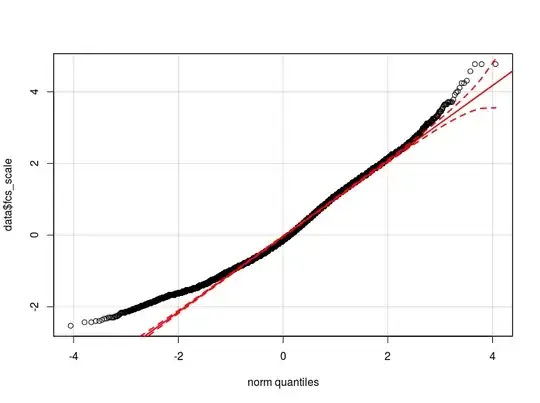
I think that the problem is in the distribution of my dependent variable. As far as I have been able to check, the normal distribution is the best approximation of the actual distribution of my data (maybe not close enough) I attach here two qq-plot respectively with the dependent variable normalized and logarithmic transformed (in red the Normal theoretical quantiles).
- Given the distribution of my variable, may the heteroskedasticity be caused by the non-normality in the dependent variable (which causes non-normality in the model's errors?)
- Should I transform the dependent variable? Should I apply a glm model? -I have tried with glm but nothing has changed in terms of BP-test output.
Do I have more efficient ways to control for the between group variation and get rid of heteroskedasticity (random intercept mixed model)?
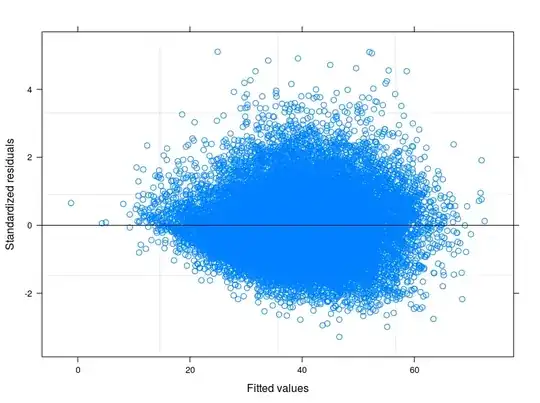
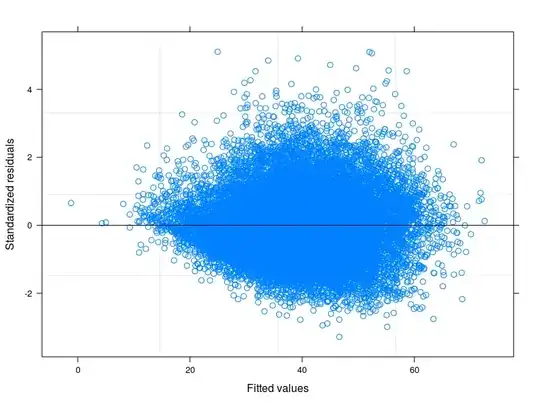
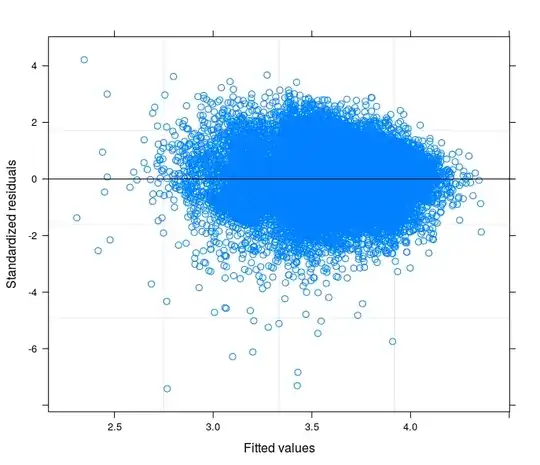
EDIT 1: I have checked in the technical manual of the Food Consumption Score, and it is reported that usually the indicator follows a "close to normal" distribution. Indeed the Shapiro-Wilk Test rejects the null hypothesis that my variable is normally distributed ( I have been able to run the test on the first 5000 obs). What I can see from the plot of the fitted against the residuals is that for lower values of fitted the variability in the errors decreases. I attach the plot here below. The plot comes from a Linear Mixed Model, to be precise a Random Intercept Model taking into account 398 different groups (Inter correlation coefficient = 0.32, groups' mean reliances not less than 0.80). Althought I have taken into account for between-group variability, heteroskedasticity is still there.
I have also runned diverse quantile regressions. I was particularly interested in the regression on the 0.25 quantile, however no improvements in terms of equal variance of the errors.
I am now thinking to account for the diversity between quantiles and groups (geographical regions) at the same time by fitting a Random Intercept Quantile Regression. May be a good idea?
Further, the Poisson distribution looks like following the trend of my data, even if for low values of the variable it wanders a bit (a bit less than the normal). However, the problem is that fitting glm of the Poisson family requires postive integers, my variable is positive but does not have exclusively integers. I thus discarded the glm (or glmm) option.
Most of your suggestions go in the direction of robust estimators. However, I think is just one of the solutions. Understanding the reason of heteroskedasticity in my data would improve the understanding of the relation I want to model. Here is clear that something is going on on the bottom of the error distribution - look at this qqplot of residuals from an OLS specification.
Any idea comes into your mind on how to further deal with this problem? Should I investigate more with quantile regressions?
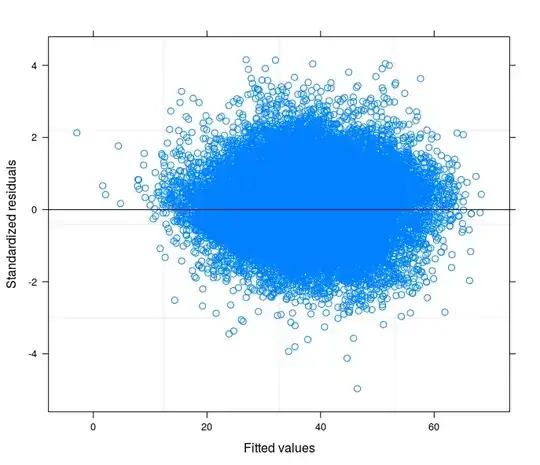
Following your suggestions I have finally runned a random intercept model tring to relate the technical problem to the theory of my field of study. I have found a variable that if included in the random part of the model makes the error terms going to homoskedasticity. Here I post 3 plots:
- The first is computed from a Random Intercept Model with 34 groups (provinces)
- The second comes from a Random Coefficient Model with 34 groups (provinces)
- Finally the third is the result of the estimation of a Random Coefficient Model with 398 groups (districts).
May I safely say that I am controlling for heteroskedasticity in the last specification?