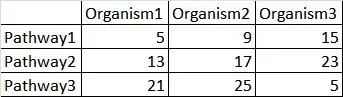
I have the counts of genes in particular pathways from three organisms. How to statistically prove that few counts are significantly high or low as compared to another organism.
Is it possible to calulate p value for particular count compared between organisms.
e.g. how to check in row 3 whether value, 5 (Org3) is significantly different (lower) from 25 and 21 (i.e. from org1 and org2) or not?
Please suggest which test is suitable and how to go about.
UPDATED ONE POSSIBLE ANSWER MYSELF.