I'm trying to classify (LDA) few samples (n=12) in a high dimensional feature space (p=24) into 3 classes.
First I reduced the dimension of my initial dataset with a PCA, keeping only the first two Eigen vectors. Update: turns out, I was actually using all 11 PCs for the LDA. Then I had a look at the projection of my n x 2 n x 11 dataset in the LDA space (1st vs 2nd Eigen vector) and I obtained the following:
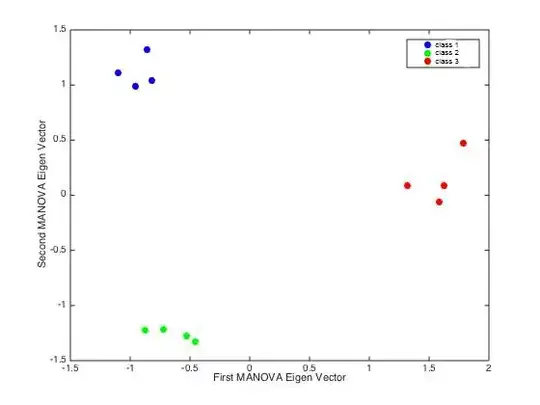
I was quite happy because the LDA found a strong separation between the 3 classes.
So I tried a leave-one-out cross validation to evaluate the LDA. I trained the classifier with 11 samples and tested it with the last one, and looped around.
The problem is the classifier performs at chance level (30% success rate).
I noticed that the LDA space changes drastically between each iteration, depending on the 11 samples used to compute it. Moreover, when I project the tested sample in the corresponding LDA space, it falls quite far away from what should be its group, explaining the poor success rate.

My questions are: is it normal that such a (visually) nice separation between classes leads to such a poor classification? Is it due to the small number of samples? Is there anything I can do to improve the situation?