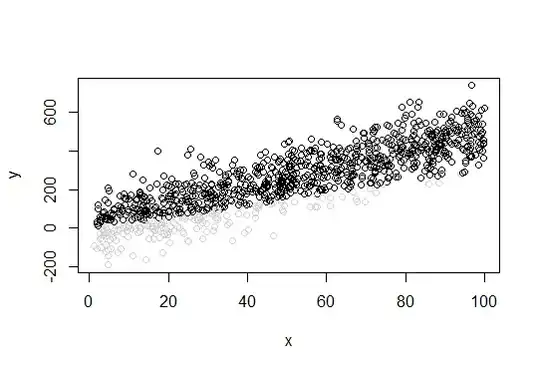
My data looks like this
n <- 1000
x <- runif(n, min=1, max=100)
y <- rnorm(n, mean=2+5*x, sd=90)
treshold <- 3
idx <- which((y/x)>treshold)
dat_trunc <- data.frame(x=x[idx],y=y[idx])
dat_full <- data.frame(x=x,y=y)
clr <- rep("lightgray", n)
clr[idx] <- "black"
plot(x,y, col=clr)
The gray data points are truncated.
Now I wish to recover the parameters of the distribution using the incomplete data.
Any hints?
EDIT: simplified the problem