I want to use normalisation technique that has assumption of residuals' normality (GLMs), but my data is $\sim$Negative Binomial.
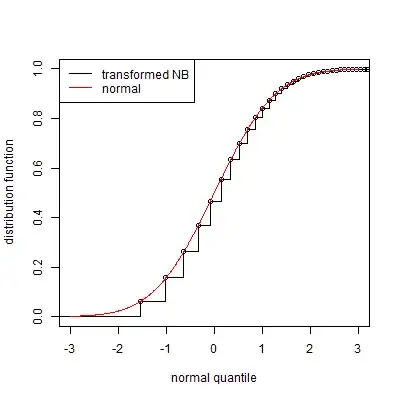
Can I map values from NB distributed distribution to Normal distribution, using probabilities and cumulative distribution function? I.e., I have a point $a$ and NB CDF says that 45% of points are less than $a$, can I map it to the point with the same property in Normal, apply normalisation and then apply reverse transformation to normalised data?
Switch to NB GLM is not feasible (the normalisation technique is really complicated, but effective, so it is not possible to quickly modify it for NB case). Authors of normalisation procedure recommend just to use variance-stabilization transformation (such as log or Anscombe), but I am not sure if it will be enough.
UPD: Data is experimental and there are a lot of data points. Bad data points (with small $mu$ and large variance, that can create problems for left tail) can be removed. What I really want to be able to do after this transformation: 1) remove batch effects using PCA or similar methods, 2) compare datapoints between samples.
UPD about PEER: this is the paper. The important thing is an equation 1). Let me explain. For each datapoint in sample $i$ and row $j$ I have a whole number $x_{ij}$ from $NB$ distribution. I need to transform $NB$-distributed data with the function $f$ to the data that can be modelled as: $RV(f(x_{ij})| conditions) \sim \mathcal N(different\_noise|conditions)$. So I need to find mapping from $NB$ distribution to somehow normal (with additional noise, etc).
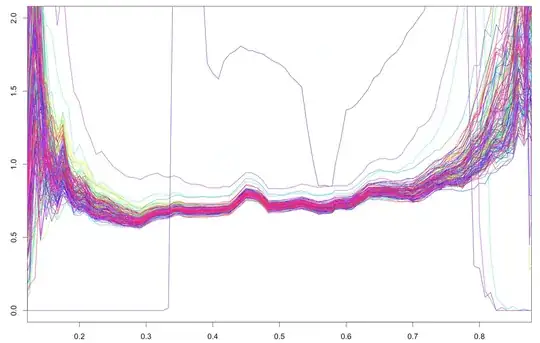
UPD about different variances across the samples: on $x$-axis is GC content, on $y$-axis: robust standard deviation of logarithms across all fragments with specified GC content, different plots - different samples.