I am trying to interpret the three boxplots below in light of another plot before finalizing my glmer models (GLMM) in R.
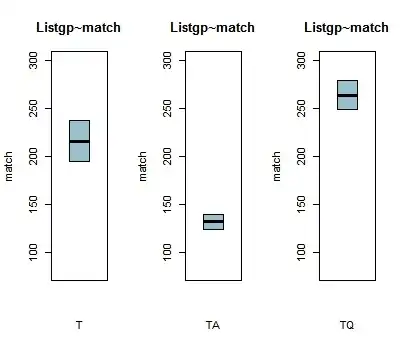
The boxplots below show the relationship between a listener's group (T, TA and TQ) and whether the listeners would match a selection or mismatch it to a predicted response (the DV match is binary: match/mismatch).
Both Listgp and match are factors; thus, I first used the xtabs function, then plotted the boxplots.
My basic model is as follows.
glmer<-(match~Listgp + (1|stimulus) + (1|listener), data= msba, family= "binomial")
Now, first, I notice that there are no whiskers in the upper and lower quantiles. I read somewhere that this means that the bottom and the top of the box would be at the exact position of the lower and upper quantiles. Is this the case here too?
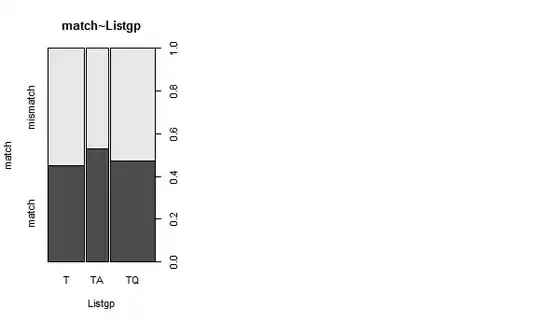
Second, I am trying to interpret these boxplots in light of the plot that follows them which is straightforward for me.
In the boxplots, the range of variance for the T group is bigger than the TQ or the TA. In addition, the TQ group seems (according to the boxplots) to be matching the patterns more, followed by the T and finally the TA.
On the other hand, according to the plot, the TA group matches the patterns more than the TA and the TQ. Basically, the TA mismatches the patterns more than the two other groups.
So my question is, do the two plots give the same or different information and is my above interpretation correct?
The first 6 lines of the data are as follows.
head(msba)
list.Listgp cond.by gender age level.of.ed. Avg.skills.T read.A comp.A speak.A
1 T1 T Qualtrics Male 29 college 10 NA NA NA
2 T1 T Qualtrics Male 29 college 10 NA NA NA
3 T1 T Qualtrics Male 29 college 10 NA NA NA
4 T1 T Qualtrics Male 29 college 10 NA NA NA
5 T1 T Qualtrics Male 29 college 10 NA NA NA
6 T1 T Qualtrics Male 29 college 10 NA NA NA
writ.A st. match st.length st.context st.nature freq.
1 NA man match short plain Real 7.80
2 NA Dhabb mismatch short emphatic Real 50.02
3 NA qad mismatch short q Real 96.57
4 NA ?an match short pharyngeal Real 15.78
5 NA min mismatch short plain Real 59.07
6 NA dhidd mismatch short emphatic Real 54.50
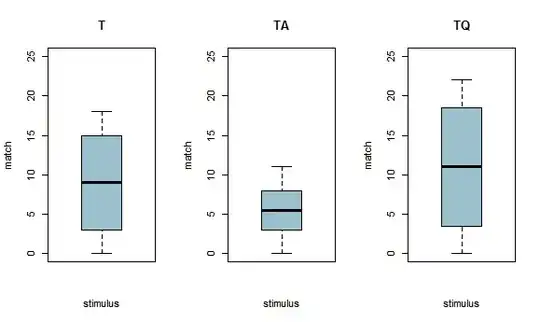
I plotted other boxplots but this time with the variable stimulus (which is also is a factor). These ones have whiskers! (Because I don't have enough reputation points, I will have to remove the 2nd plot so that I can paste the boxplots with stimulus)
I used these codes for the xtabs.
monosbaT.xtabs <- xtabs(~ match + st. , data = monosbaT)
monosbaTA.xtabs <- xtabs(~ match + st., data = monosbaTA)
monosbaTQ.xtabs <- xtabs(~ match + st., data = monosbaTQ)
Then, in the lattice package I plotted them using these codes.
par(mfrow = c(1,3))
boxplot (monosbaT.xtabs, main = "T", legend.text= c("match", "mismatch"), xlab= "stimulus", ylab= "match", col= "lightblue3", ylim=c(0, 25), breaks=seq(0, 25, 8))
boxplot (monosbaTA.xtabs, main = "TA", legend.text= c("match", "mismatch"), xlab= "stimulus", ylab= "match", col= "lightblue3", ylim=c(0, 25), breaks=seq(0, 25, 8))
boxplot (monosbaTQ.xtabs, main = "TQ", legend.text= c("match", "mismatch"), xlab= "stimulus", ylab= "match", col= "lightblue3", ylim=c(0, 25), breaks=seq(0, 25, 8))
par(mfrow =c(1,1))
My dataset has 1224 obs. of 17 variables.