In order to calculate the mode of a continious distribution in python I suggest three options. Scipy have scipy.stats.mode(data)[0] but it's not exact. For the three options after we need to get an approximation of de PDF function of the data. An excellent method is Gaussian Kernel
Density Estimation(WIKIPEDIA). With scipy we can use distribution = scipy.stats.gaussian_kde(data) and get pdf value of the dataset with distribution.pdf(x)[0]. The three methods search the max value of the distribution and get the preimage of the value in the the domian. The first use scipy minimize method, the second use max() native function, and the third SHGO scipy method. Here and imgage of comparison:
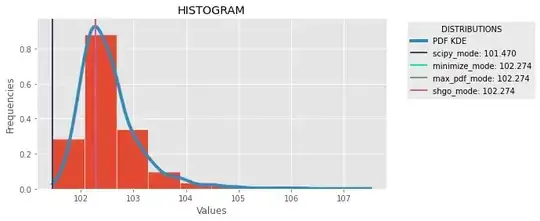 And the complete code:
And the complete code:
import numpy as np
import scipy.stats
import scipy.optimize
import matplotlib as mpl
import matplotlib.pyplot as plt
import random
mpl.style.use("ggplot")
def plot_histogram(data, distribution, modes):
plt.figure(figsize=(8, 4))
plt.hist(data, density=True, ec='white')
plt.title('HISTOGRAM')
plt.xlabel('Values')
plt.ylabel('Frequencies')
x_plot = np.linspace(min(data), max(data), 1000)
y_plot = distribution.pdf(x_plot)
plt.plot(x_plot, y_plot, linewidth=4, label="PDF KDE")
for name, mode in modes.items():
plt.axvline(mode, linewidth=2, label=name+": "+str(mode)[:7], color=(random.uniform(0, 1), random.uniform(0, 1), random.uniform(0, 1)))
plt.legend(title='DISTRIBUTIONS', bbox_to_anchor=(1.05, 1), loc='upper left')
plt.show()
## SCIPY MODE
def calc_scipy_mode(data):
return scipy.stats.mode(data)[0]
## METHOD 1: MAXIMIZE PDF SCIPY MINIMIZE
def calc_minimize_mode(data, distribution):
def objective(x):
return 1/distribution.pdf(x)[0]
bnds = [(min(data), max(data))]
solution = scipy.optimize.minimize(objective, [1], bounds = bnds)
return solution.x[0]
## METHOD 2: MAXIMIZE PDF AND GET PREIMAGE
def calc_max_pdf_mode(data, distribution):
x_domain = np.linspace(min(data), max(data), 1000)
y_pdf = distribution.pdf(x_domain)
i = np.argmax(y_pdf)
return x_domain[i]
## METHOD 3: ## METHOD 3: MAXIMIZE PDF SCIPY SHGO
def calc_shgo_mode(data, distribution):
def objective(x):
return 1/distribution.pdf(x)[0]
bnds = [[min(data), max(data)]]
solution = scipy.optimize.shgo(objective, bounds= bnds, n=100*len(data))
return solution.x[0]
def calculate_mode(data):
## KDE
distribution = scipy.stats.gaussian_kde(data)
scipy_mode = calc_scipy_mode(data)[0]
minimize_mode = calc_minimize_mode(data, distribution)
max_pdf_mode = calc_max_pdf_mode(data, distribution)
shgo_mode = calc_shgo_mode(data, distribution)
modes = {
"scipy_mode": scipy_mode,
"minimize_mode": minimize_mode,
"max_pdf_mode": max_pdf_mode,
"shgo_mode": shgo_mode
}
plot_histogram(data, distribution, modes)
if __name__ == "__main__":
data = [d1, d2, d3, ..........]
calculate_mode(data)
