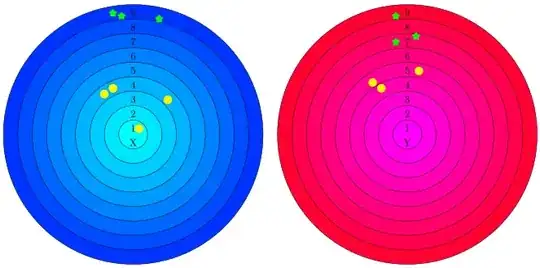
I would like to evaluate a similarity between two objects X and Y by comparing a neighbourhood in which they're located.
I construct two sets of nine concentric and equidistant circles with centers in X and Y respectively (nine circles, but utterly not Dante's infernal ;-D )
This considered objects X and Y are surrounded by many other objects of different types. I consider all objects located only in this nine rings. In the figure above, we have two types of objects: yellow balls and green stars.
I'm counting occurrences of those surrounding objects, separately of each type and in each ring, creating two ordinary vectors. (Below, I've splitted them by type only for clarity, but generally that are two vectors: one for X and one for Y.) In our example: $X_{YellowBalls} = [1, 0, 0, 3, 0, \dots, 0]$, $X_{GreenStars} = [0, \dots, 0, 0, 3]$, $Y_{YellowBalls} = [0, 0, 0, 1, 2, \dots, 0]$, $X_{GreenStars} = [0, \dots, 2, 0, 1]$.
Now, I'm trying to infere something about similarity of X and Y on the basis of these two vecors using many vector similarity measure, which are mainly used in text mining for document comparision.
The main problem lies here: This off-the-shelf methods are prone to differentiate a way too much vector entries obtained by counting in consecutive rings, e.g. for X three yellow balls are gathered in 4th ring whereas for Y only one yellow ball is located in 4th ring and two are placed in adjoining 5th ring. They are in pronounced proximity, but they fall into different rings only by hapenstance, because they ran into a border.
A very similar partitions concers green stars, 3 in 9th against 1 in 9th and 2 in 7th), but they're not in neighbouring rings, so that's completly different story. In my opinion, a proper method of comparision should be able to catch such nicity and threat these two situations in a sundry manner.
The ultimate question is: are there any methods for vectors similarity measures that will be more aware to neighbouring positions in vectors.
So far, I've tried several adjustments, but unfortunately, none of my ideas improved it to a satisfactory extent.
Does anyone know any publications that cope with similar problem?