What is the standard method for generating a 95% predicton interval (not confidence) for a linear regression given heteroscedastic data?
Let me be more specific with an example:
dummySamples <- function(n, slope, intercept, slopeVar){
x = runif(n)
y = slope*x+intercept+rnorm(n, mean=0, sd=slopeVar*x)
return(data.frame(x=x,y=y))
}
> myDF <- dummySamples(20000,3,0,5)
> plot(myDF$x, myDF$y)
> t = lm(y~x, data=myDF)
> summary(t)
Call:
lm(formula = y ~ x, data = myDF)
Residuals:
Min 1Q Median 3Q Max
-17.8120 -1.3153 -0.0059 1.3243 18.1977
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.003637 0.040708 0.089 0.929
x 2.927918 0.070574 41.487 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 2.878 on 19998 degrees of freedom
Multiple R-squared: 0.07925, Adjusted R-squared: 0.0792
F-statistic: 1721 on 1 and 19998 DF, p-value: < 2.2e-16
> newdata = data.frame(x=seq(0,1,0.01))
> p1 = predict(t, newdata, interval = 'predict')
> a <- ggplot()
> a <- a + geom_point(data=myDF, aes(x=x,y=y), shape=1)
> a <- a + geom_abline(intercept=t$coefficients[1], slope=t$coefficients[2])
> a <- a + geom_abline(intercept=t$coefficients[1], slope=t$coefficients[2], color='blue')
> a <- ggplot()
> a <- a + geom_point(data=myDF, aes(x=x,y=y), shape=1)
> a <- a + geom_abline(intercept=t$coefficients[1], slope=t$coefficients[2], color='blue')
> newdata$lwr = p1[,c("lwr")]
> newdata$upr = p1[,c("upr")]
> a <- a + geom_ribbon(data=newdata, aes(x=x,ymin=lwr, ymax=upr), fill='yellow', alpha=0.3)
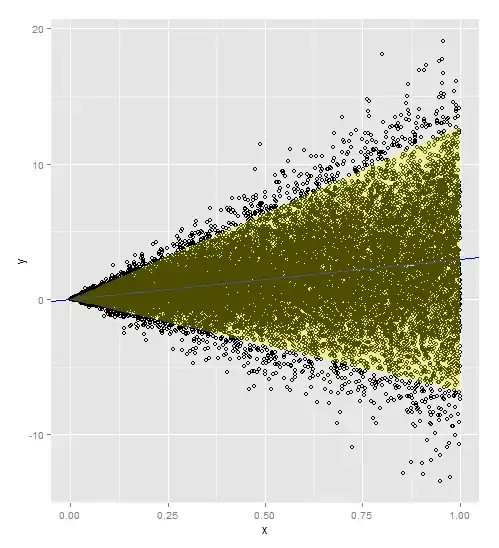
> a
Since I simulated a lot of points from a known function, I can see that the 95% prediction interval underestimates (too narrow) where the variance is larger and overestimates (too wide) where the variance is smaller. Is there a standard method for generating a 95% prediction interval that better fits the heteroscedastic data?
I will not have this many points in reality, I just wanted to clearly demonstrate my question.
Thanks.
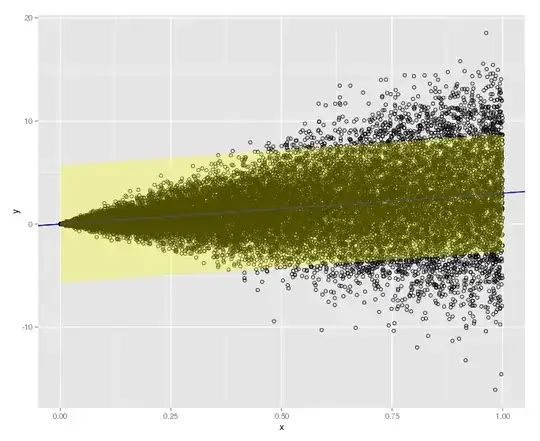
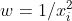 as your weight. This weights your variance by your x value, so that there's a proportional relationship.
as your weight. This weights your variance by your x value, so that there's a proportional relationship.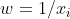 weight, but your nonconstant variance is more extreme, so a
weight, but your nonconstant variance is more extreme, so a