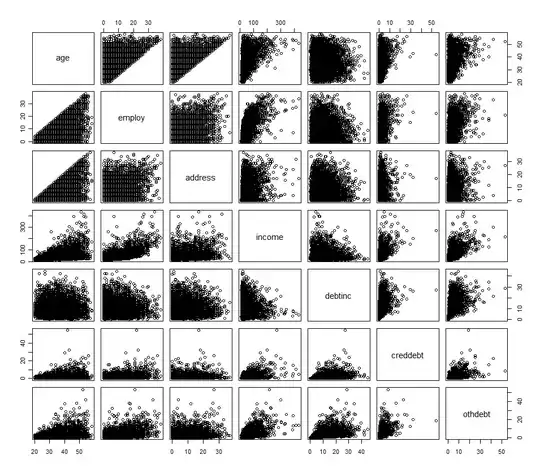
Even though you say that the geometry of this is fairly clear to you, I think it is a good idea to review it. I made this back of an envelope sketch:
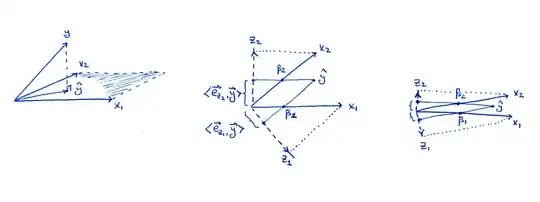
Left subplot is the same figure as in the book: consider two predictors $x_1$ and $x_2$; as vectors, $\mathbf x_1$ and $\mathbf x_2$ span a plane in the $n$-dimensional space, and $\mathbf y$ is being projected onto this plane resulting in the $\hat {\mathbf y}$.
Middle subplot shows the $X$ plane in the case when $\mathbf x_1$ and $\mathbf x_2$ are not orthogonal, but both have unit length. The regression coefficients $\beta_1$ and $\beta_2$ can be obtained by a non-orthogonal projection of $\hat{\mathbf y}$ onto $\mathbf x_1$ and $\mathbf x_2$: that should be pretty clear from the picture. But what happens when we follow the orthogonalization route?
The two orthogonalized vectors $\mathbf z_1$ and $\mathbf z_2$ from Algorithm 3.1 are also shown on the figure. Note that each of them is obtained via a separate Gram-Schmidt orthogonalization procedure (separate run of Algorithm 3.1): $\mathbf z_1$ is the residual of $\mathbf x_1$ when regressed on $\mathbf x_2$ ans $\mathbf z_2$ is the residual of $\mathbf x_2$ when regressed on $\mathbf x_1$. Therefore $\mathbf z_1$ and $\mathbf z_2$ are orthogonal to $\mathbf x_2$ and $\mathbf x_1$ respectively, and their lengths are less than $1$. This is crucial.
As stated in the book, the regression coefficient $\beta_i$ can be obtained as $$\beta_i = \frac{\mathbf z_i \cdot \mathbf y}{\|\mathbf z_i\|^2} =\frac{\mathbf e_{\mathbf z_i} \cdot \mathbf y}{\|\mathbf z_i\|},$$ where $\mathbf e_{\mathbf z_{i}}$ denotes a unit vector in the direction of $\mathbf z_i$. When I project $\hat{\mathbf y}$ onto $\mathbf z_i$ on my drawing, the length of the projection (shown on the figure) is the nominator of this fraction. To get the actual $\beta_i$ value, one needs to divide by the length of $\mathbf z_i$ which is smaller than $1$, i.e. the $\beta_i$ will be larger than the length of the projection.
Now consider what happens in the extreme case of very high correlation (right subplot). Both $\beta_i$ are sizeable, but both $\mathbf z_i$ vectors are tiny, and the projections of $\hat{\mathbf y}$ onto the directions of $\mathbf z_i$ will also be tiny; this is I think what is ultimately worrying you. However, to get $\beta_i$ values, we will have to rescale these projections by inverse lengths of $\mathbf z_i$, obtaining the correct values.
Following the Gram-Schmidt procedure, the residual of X1 or X2 on the other covariates (in this case, just each other) effectively remove the common variance between them (this may be where I am misunderstanding), but surely doing so removes the common element that manages to explain the relationship with Y?
To repeat: yes, the "common variance" is almost (but not entirely) "removed" from the residuals -- that's why projections on $\mathbf z_1$ and $\mathbf z_2$ will be so short. However, the Gram-Schmidt procedure can account for it by normalizing by the lengths of $\mathbf z_1$ and $\mathbf z_2$; the lengths are inversely related to the correlation between $\mathbf x_1$ and $\mathbf x_2$, so in the end the balance gets restored.
Update 1
Following the discussion with @mpiktas in the comments: the above description is not how Gram-Schmidt procedure would usually be applied to compute regression coefficients. Instead of running Algorithm 3.1 many times (each time rearranging the sequence of predictors), one can obtain all regression coefficients from the single run. This is noted in Hastie et al. on the next page (page 55) and is the content of Exercise 3.4. But as I understood OP's question, it referred to the multiple-runs approach (that yields explicit formulas for $\beta_i$).
Update 2
In reply to OP's comment:
I am trying to understand how 'common explanatory power' of a (sub)set of covariates is 'spread between' the coefficient estimates of those covariates. I think the explanation lies somewhere between the geometric illustration you have provided and mpiktas point about how the coefficients should sum to the regression coefficient of the common factor
I think if you are trying to understand how the "shared part" of the predictors is being represented in the regression coefficients, then you do not need to think about Gram-Schmidt at all. Yes, it will be "spread out" between the predictors. Perhaps a more useful way to think about it is in terms of transforming the predictors with PCA to get orthogonal predictors. In your example there will be a large first principal component with almost equal weights for $x_1$ and $x_2$. So the corresponding regression coefficient will have to be "split" between $x_1$ and $x_2$ in equal proportions. The second principal component will be small and $\mathbf y$ will be almost orthogonal to it.
In my answer above I assumed that you are specifically confused about Gram-Schmidt procedure and the resulting formula for $\beta_i$ in terms of $z_i$.