Yes, even a single A-outlier (sample of class A) placed in the middle of many B examples (in a feature space) would affect the structure of the forest. The trained forest model will predict new samples as A, when these are placed very close to the A-outlier. But the density of neighboring B examples will decrease size of this "predict A"-island in the "predict B"-ocean. But the "predict A"-island will not disappear.
For noisy classification, e.g. Contraceptive Method Choice, the default random forest can be improved by lowering tried varaibles in each split(mtry) and bootstrap sample size(sampsize). If sampsize is e.g. 40% of training size, then any 'single sample prediction island' will completely drown if surrounded by only counter examples, as it only will be present in 40% of the trees.
EDIT: If sample replacement is true, then more like 33% of trees.
mean(replicate(1000,length(unique(sample(1:1000,400,rep=T)))))
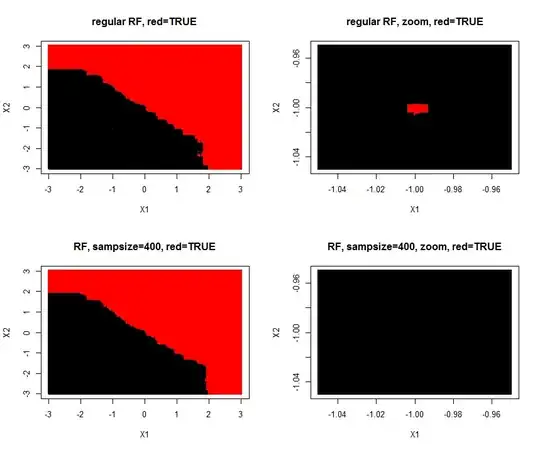
I made a simulation of the problem (A=TRUE,B=FALSE) where one (A/TRUE)sample is injected within many (B/FALSE) samples. Hereby is created a tiny A-island in the B ocean. The area of the A-island is so small, it has no influence on the overall prediction performance. Lowering sample size makes the island disappear.
1000 samples with two features $X_1$ and $X_2$ are of class "true/A" if $y_i = X_1 + X_2 >= 0$. Features are drawn from $N(0,1)$
library(randomForest)
par(mfrow=c(2,2))
set.seed(1)
#make data
X = data.frame(replicate(2,rnorm(1000)))
y = factor(apply(X,1,sum) >=0) #create 2D class problem
X[1,] = c(-1,-1); y[1]='TRUE' #insert one TRUE outlier inside 'FALSE-land'
#train default forest
rf = randomForest(X,y)
#make test grid(250X250) from -3 to 3,
Xtest = expand.grid(replicate(2,seq(-3,3,le=250),simplify = FALSE))
Xtest[1,] = c(-1,-1) #insert the exact same coordinate of train outlier
Xtest = data.frame(Xtest); names(Xtest) = c("X1","X2")
plot(Xtest,col=predict(rf,Xtest),pch=15,cex=0.5,main="regular RF, red=TRUE")
#zoom in on area surrouding outlier
Xtest = expand.grid(replicate(2,seq(-1.05,-.95,le=250),simplify = FALSE))
Xtest = data.frame(Xtest); names(Xtest) = c("X1","X2")
plot(Xtest,col=predict(rf,Xtest),pch=15,cex=0.5,main= "regular RF, zoom, red=TRUE")
#train extra robust RF
rf = randomForest(X,y,sampsize = 400)
Xtest = expand.grid(replicate(2,seq(-3,3,le=250),simplify = FALSE))
Xtest[1,] = c(-1,-1)
Xtest = data.frame(Xtest); names(Xtest) = c("X1","X2")
plot(Xtest,col=predict(rf,Xtest),pch=15,cex=0.5,main="RF, sampsize=400, red=TRUE")
Xtest = expand.grid(replicate(2,seq(-1.05,-.95,le=250),simplify = FALSE))
Xtest = data.frame(Xtest); names(Xtest) = c("X1","X2")
plot(Xtest,col=predict(rf,Xtest),pch=15,cex=0.5,main="RF, sampsize=400, zoom, red=TRUE")
