I am working with camera-trap data on mammals. My data looks like this:
Zone Point Phase SurveyLength ProjectDay Species1 Species2
A A1 Before 21 1 0 0
A A1 Before 21 2 1 0
A A1 Before 21 3 0 1
...
B B1 Before 21 1 2 0
...
B B2 After 21 1 0 0
...
B B3 After 21 1 0 1
...
There are ten species in total. I want to compare detection rate between zones and phases, as this is a BACI experiment. Detection rate is measured as the number of observations of a given species divided by effort; effort = camera days. so essentially, detection rate = average for the entire dataset for each species).
I have separated the data by phase, then analyze using a glm. However, even with the most abundant species, the model fails the goodness of fit test. Here is an example of the code I'm using:
before <- subset(Counts, Counts$Phase=="Before")
ZoneB <- factor(before$Zone)
DurationDaysB <- as.numeric(before$SurveyLength)
glmB <-glm(Species1~ ZoneB, family=poisson(link=log), offset=log(DurationDaysB), data=before)
count.covB <- cbind(ZoneB)
chsq<-sum(residuals(glmB, type = 'pearson') ^ 2)
gofB <- POIS_GOF(mu = glmB$coefficients, sigma = vcov(glmB),
sims = 1000, chsq.obs=chsq, count.cov = count.covB,
offset.count = log(DurationDaysB))
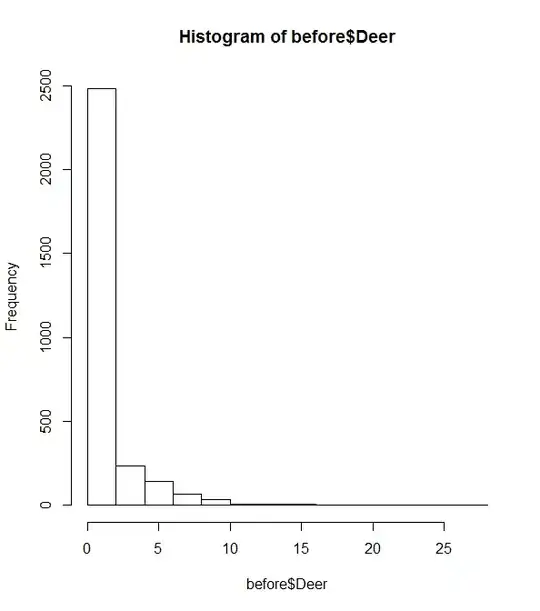
There are a LOT of zero's in my data (histogram included below). After trying the species with the top three detection rates, p=0 in every case, indicating lack of fit with the model.
I've read on the UCLA website about a way to compensate for overdispersion by scaling the data in STATA, but I have not found a way to do this in R. I also tried using the "quasipoisson" distribution, which should allow for overdispersion in models where the dispersion parameter is not fixed. I have not fixed this in my model, but the quasipoisson model returns the same results as the poisson model.
Is there another way I can compensate for overdispersion in my data? Or is there a different model which I should use for this type of data?
I also ran the analysis using an anova followed by a Tukey test from the raw data (as shown below), but then remembered that I would need to use a Poisson distribution for this data. While we're here, I want to verify that this is an incorrect way to analyze the data - thoughts?
species1countB <- aov(before$Species1~before$Zone)
Thank you in advance for your help.