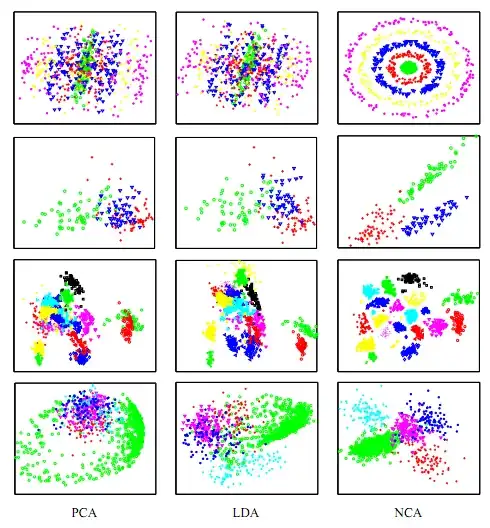
So let's say I have a bunch of data points in R^n, where n is pretty big (like, 50). I know this data falls into 3 clusters, and I know which cluster each data point is a part of. All I want to do is visualize these clusters in 2D in such a way as to maximize the visual between-clusters separation that I see, with the goal being to prove that the clusters are easily separable based on the location of the data point in R^n alone.
The way I've been going about this up until now involves doing a PCA transform on the data points and then visualizing pairs of PCs at random until I find one where the clusters appear to be pretty cleanly separated. This approach seems pretty ad hoc though, and it seems like there should be an easy way to find a PCA-style rotation of the data that, instead of maximizing overall variance, maximizes inter-cluster separation.
Is there a standard technique out there that does this? If not, any ideas about how to create such a transformation?