I have previously posted about the data I am using in this logistic regression here - Skewed Distributions for Logistic Regression
I have built a logistic regression model and tested using the rms package. The regression model included the following variables:
Yeardecimal - Date of procedure (expressed as decimal of year) = 1994-2013
inctoCran - Time from head injury to craniotomy in minutes = 0-2880 (After 2880 minutes is defined as a separate diagnosis)
Age - Age of patient = 16.0-101.5
rcteyemi - Pupil reactivity = NA / Both unreactive = O, 1 reactive = 1, both reactive = 2
GCS - Glasgow Coma Scale = 3-15
ISS - Injury Severity Score = 16-75
Other - dummy for presence (1) or absence (0) of other trauma
SDH Severity - Score for severity of subdural haematoma (4 or 5)
Mechanism - Mechanism of injury = Fall <2m, Fall >2m, Shooting/stabbing, RTC (Road Traffic Collision), Other
neuroFirst2 - Location of first admission (Neurosurgical Unit) = NSU vs. Non-NSU
Sex - Gender of patient = Male or Female
Weekday - Day of the week of injury
Multiple imputation was performed due to missingness in the data (GCS 14% missing, rcteyemi 46% missing).
The results of the final model are as follows:
fit.mult.impute(formula = Survive ~ rcs(Yeardecimal, 4) + rcs(inctoCran) +
rcs(Age) + rcteyemi + rcs(GCS, 4) + rcs(ISS, 3) + Other +
SDH.Severity * Other + rcs(ISS, 3) * SDH.Severity + Mechanism +
neuroFirst2 + Sex + Weekday, fitter = lrm, xtrans = a, data = ASDH_Paper1.1)
Model Likelihood Discrimination Rank Discrim.
Ratio Test Indexes Indexes
Obs 2498 LR chi2 760.36 R2 0.373 C 0.822
0 737 d.f. 35 g 1.643 Dxy 0.644
1 1761 Pr(> chi2) <0.0001 gr 5.173 gamma 0.645
max |deriv| 1e-04 gp 0.269 tau-a 0.268
Brier 0.146
Coef S.E. Wald Z Pr(>|Z|)
Intercept -127.7493 75.1796 -1.70 0.0893
Yeardecimal 0.0654 0.0376 1.74 0.0815
Yeardecimal' -0.0340 0.0557 -0.61 0.5419
Yeardecimal'' 0.4371 0.7780 0.56 0.5743
inctoCran 0.0006 0.0020 0.31 0.7585
inctoCran' -0.0652 0.1590 -0.41 0.6818
inctoCran'' 0.1507 0.3393 0.44 0.6570
inctoCran''' -0.0981 0.2003 -0.49 0.6243
Age -0.0158 0.0229 -0.69 0.4892
Age' 0.0264 0.1250 0.21 0.8328
Age'' -0.2912 0.4308 -0.68 0.4991
Age''' 0.6521 0.5444 1.20 0.2310
rcteyemi=1 0.7157 0.2121 3.37 0.0007
rcteyemi=2 2.4627 0.2123 11.60 <0.0001
GCS 0.0732 0.0809 0.90 0.3655
GCS' 0.0462 0.3506 0.13 0.8952
GCS'' -0.1736 0.7046 -0.25 0.8054
ISS -0.1733 0.0331 -5.24 <0.0001
ISS' 0.1127 0.0331 3.41 0.0007
Other=1 0.6694 0.2461 2.72 0.0065
SDH.Severity=5 -1.1855 5.3867 -0.22 0.8258
Mechanism=Fall > 2m 0.0483 0.1588 0.30 0.7612
Mechanism=Other 0.5464 0.1922 2.84 0.0045
Mechanism=RTC 0.1914 0.1744 1.10 0.2726
Mechanism=Shooting / Stabbing 1.1738 1.1886 0.99 0.3234
neuroFirst2=NSU -0.2079 0.1274 -1.63 0.1027
Sex=Male -0.2042 0.1320 -1.55 0.1219
Weekday=Monday 0.0846 0.2031 0.42 0.6770
Weekday=Saturday 0.1279 0.1917 0.67 0.5048
Weekday=Sunday 0.2215 0.1928 1.15 0.2506
Weekday=Thursday 0.2590 0.2147 1.21 0.2276
Weekday=Tuesday 0.0696 0.2159 0.32 0.7472
Weekday=Wednesday -0.1883 0.2113 -0.89 0.3727
Other=1 * SDH.Severity=5 -0.8443 0.4738 -1.78 0.0748
ISS * SDH.Severity=5 0.0533 0.2256 0.24 0.8131
ISS' * SDH.Severity=5 -0.0129 0.1930 -0.07 0.9465
p-values for each variable were identified using anova():
Wald Statistics Response: Survive
Factor Chi-Square d.f. P
Yeardecimal 16.60 3 0.0009
Nonlinear 0.37 2 0.8297
inctoCran 3.31 4 0.5075
Nonlinear 0.38 3 0.9453
Age 66.75 4 <.0001
Nonlinear 4.55 3 0.2077
rcteyemi 153.15 2 <.0001
GCS 11.68 3 0.0086
Nonlinear 1.06 2 0.5883
ISS (Factor+Higher Order Factors) 40.22 4 <.0001
All Interactions 3.25 2 0.1971
Nonlinear (Factor+Higher Order Factors) 11.80 2 0.0027
Other (Factor+Higher Order Factors) 7.76 2 0.0206
All Interactions 3.18 1 0.0748
SDH.Severity (Factor+Higher Order Factors) 6.54 4 0.1623
All Interactions 6.48 3 0.0906
Mechanism 9.24 4 0.0555
neuroFirst2 2.66 1 0.1027
Sex 2.39 1 0.1219
Weekday 6.08 6 0.4140
Other * SDH.Severity (Factor+Higher Order Factors) 3.18 1 0.0748
ISS * SDH.Severity (Factor+Higher Order Factors) 3.25 2 0.1971
Nonlinear 0.00 1 0.9465
Nonlinear Interaction : f(A,B) vs. AB 0.00 1 0.9465
TOTAL NONLINEAR 17.38 12 0.1357
TOTAL INTERACTION 6.48 3 0.0906
TOTAL NONLINEAR + INTERACTION 30.18 14 0.0072
TOTAL 404.15 35 <.0001
My question relates to the result of the summary function relative to the above anova results. summary() produces the following output:
Effects Response : Survive
Factor Low High Diff. Effect S.E. Lower 0.95 Upper 0.95
Yeardecimal 2004.900 2012.600 7.755 0.26 0.17 -0.08 0.60
Odds Ratio 2004.900 2012.600 7.755 1.30 NA 0.93 1.83
inctoCran 235.000 778.500 543.500 0.05 0.18 -0.30 0.39
Odds Ratio 235.000 778.500 543.500 1.05 NA 0.74 1.48
Age 33.625 64.775 31.150 -1.05 0.19 -1.41 -0.68
Odds Ratio 33.625 64.775 31.150 0.35 NA 0.24 0.51
GCS 4.000 13.000 9.000 0.57 0.19 0.19 0.94
Odds Ratio 4.000 13.000 9.000 1.76 NA 1.22 2.56
ISS 25.000 29.000 4.000 -0.20 0.36 -0.91 0.52
Odds Ratio 25.000 29.000 4.000 0.82 NA 0.40 1.67
rcteyemi - 0:2 3.000 1.000 NA -2.46 0.21 -2.88 -2.05
Odds Ratio 3.000 1.000 NA 0.09 NA 0.06 0.13
rcteyemi - 1:2 3.000 2.000 NA -1.75 0.19 -2.12 -1.37
Odds Ratio 3.000 2.000 NA 0.17 NA 0.12 0.25
Other - 1:0 1.000 2.000 NA -0.17 0.42 -1.00 0.65
Odds Ratio 1.000 2.000 NA 0.84 NA 0.37 1.92
SDH.Severity - 4:5 2.000 1.000 NA -0.13 0.17 -0.47 0.21
Odds Ratio 2.000 1.000 NA 0.88 NA 0.63 1.23
Mechanism - Fall > 2m:Fall < 2m 1.000 2.000 NA 0.05 0.16 -0.26 0.36
Odds Ratio 1.000 2.000 NA 1.05 NA 0.77 1.43
Mechanism - Other:Fall < 2m 1.000 3.000 NA 0.55 0.19 0.17 0.92
Odds Ratio 1.000 3.000 NA 1.73 NA 1.19 2.52
Mechanism - RTC:Fall < 2m 1.000 4.000 NA 0.19 0.17 -0.15 0.53
Odds Ratio 1.000 4.000 NA 1.21 NA 0.86 1.70
Mechanism - Shooting / Stabbing:Fall < 2m 1.000 5.000 NA 1.17 1.19 -1.16 3.50
Odds Ratio 1.000 5.000 NA 3.23 NA 0.31 33.23
neuroFirst2 - NSU:Non-NSU 1.000 2.000 NA -0.21 0.13 -0.46 0.04
Odds Ratio 1.000 2.000 NA 0.81 NA 0.63 1.04
Sex - Female:Male 2.000 1.000 NA 0.20 0.13 -0.05 0.46
Odds Ratio 2.000 1.000 NA 1.23 NA 0.95 1.59
Weekday - Friday:Sunday 4.000 1.000 NA -0.22 0.19 -0.60 0.16
Odds Ratio 4.000 1.000 NA 0.80 NA 0.55 1.17
Weekday - Monday:Sunday 4.000 2.000 NA -0.14 0.20 -0.53 0.25
Odds Ratio 4.000 2.000 NA 0.87 NA 0.59 1.29
Weekday - Saturday:Sunday 4.000 3.000 NA -0.09 0.18 -0.46 0.27
Odds Ratio 4.000 3.000 NA 0.91 NA 0.63 1.31
Weekday - Thursday:Sunday 4.000 5.000 NA 0.04 0.22 -0.38 0.46
Odds Ratio 4.000 5.000 NA 1.04 NA 0.68 1.58
Weekday - Tuesday:Sunday 4.000 6.000 NA -0.15 0.21 -0.56 0.26
Odds Ratio 4.000 6.000 NA 0.86 NA 0.57 1.29
Weekday - Wednesday:Sunday 4.000 7.000 NA -0.41 0.20 -0.81 -0.01
Odds Ratio 4.000 7.000 NA 0.66 NA 0.45 0.99
I am not sure exactly why I have significant p-values but an odds ratio whose confidence interval traps 1 for the following variables?
1. Yeardecimal - p-value = 0.0009, OR = 1.30 (95% CI 0.93-1.83)
2. ISS - p-value < 0.0001, OR = 0.82 (95% CI 0.40-1.67)
3. Other - p-value = 0.0206, OR = 0.84 (95% CI 0.37-1.92)
I understand that the OR is calculated for continuous variables by comparing the 75th to the 25th percentile. Non-linear restricted cubic spline modelling of Year and ISS may explain the discordance between OR and p-value but what about the binary variable Other? How could I explain this when writing up the results?
Many thanks for any help you could give with this.
Update
Below is a trajectory using plot(Predict(rcs.ASDH,Yeardecimal)) in rms:
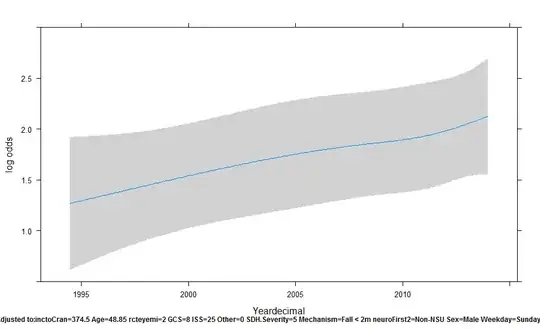
In preparing the manuscript for publication, I have converted the log odds to survival, and used ggplot(Predict(...)) to produce the following trend over time:
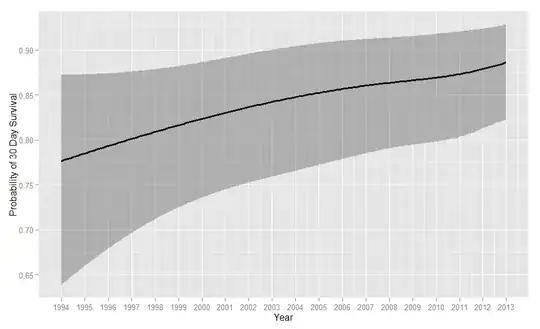
Should I remove the OR data from the results table? I am just concerned that reviewers and readers may be confused by the (to a typical medical professional) conflicting statistical results.
Update
Please see below marginal effects plot using the following code:
an<-anova(rcs.ASDH)
ggplot(Predict(rcs.ASDH,name="Yeardecimal"), anova=an, pval=TRUE)