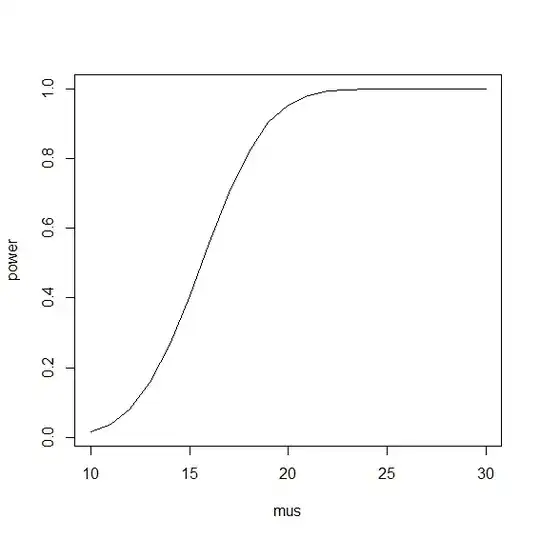
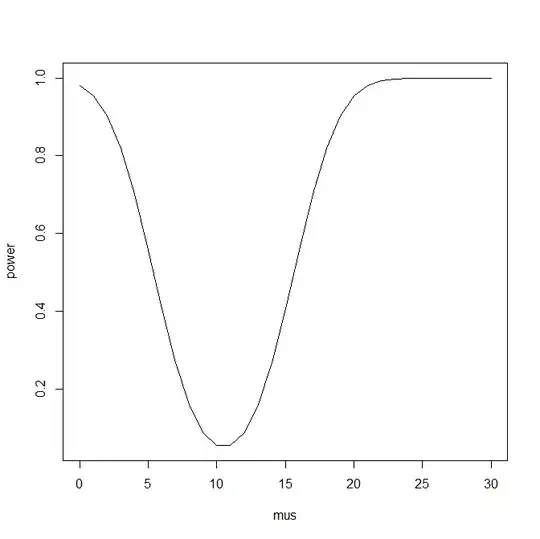
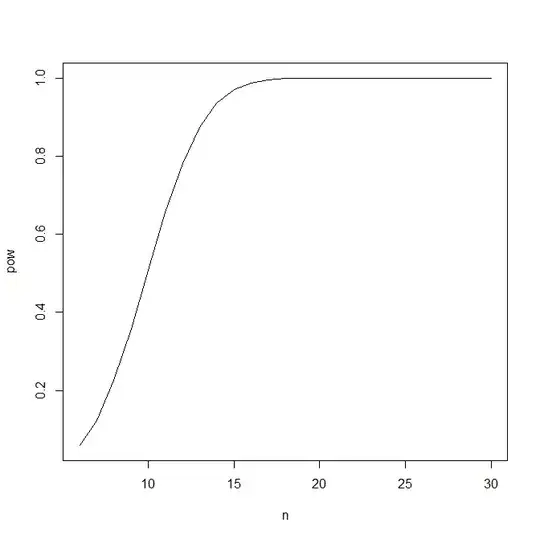
Suppose I know that the true population proportion of a mutation is p = 0.3493119. I want to know that given power = 0.8, what's the proportion of of mutation in my sample of n = 30? Here's what I have so far:
1 - cdf((x-p)/sqrt(p*(1-p)/30) = 0.8 Since p = 0.3493119, I can solve for x and I get x = 0.276055. So does this mean that the probability of having 27.6% of the sample be mutated = 0.8? Is it correct to say that?
From my experience I know that power is the probability of correctly rejecting the null. But since I already know the true population proportion of the mutation, is it still necessary to conduct a hypothesis test? I am leaning towards the negative, but without a hypothesis test, how can I calculate the power?
If I had to perform a hypothesis test to get the power...would it be something like this:
H0: p = 0.5 vs. H1: p > 0.5
power = P(Z_statistic > critical_value | p > 0.5)
power = 1 - normal_CDF(critical_value | p > 0.5)
Now I'm confused by how to deal with the p > 0.5, if it were just p = 0.5, I could've standardized the critical value so that it follows a N(0, 1) distribution.
Overall I think I just want to know the answer to the question, given power = 0.8, what is the proportion of mutation in my sample of n = 30? (true population proportion = 0.3493119).