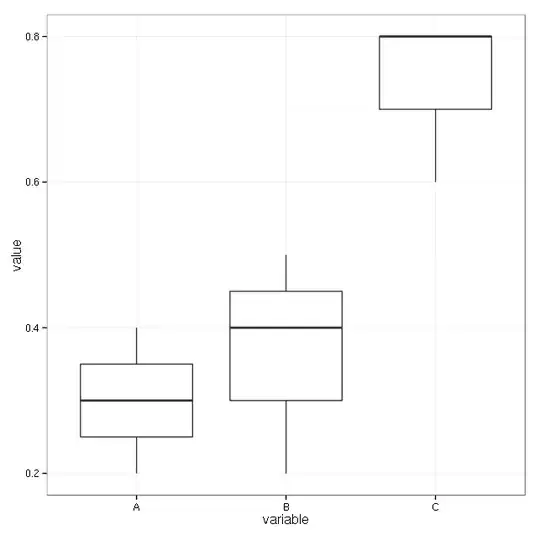
I have a problem in medicine: Each patient can have several (3) symptoms (A,B,C) of a disease at the same time. For each symptomtype we can measure a score i.e. A=0.2, B=0.45, C=0.8. The aim is that we want to conclude from the value of the score to what kind of symptom (A,B,C) the measured value belongs. So my test hypothesis is that the means of A,B,C are not different and my alternative is that they are ordered A < B < C.
Here is my table:
PatientId, A, B, C
1, 0.2, 0.5, 0.8
2, 0.3, 0.4, 0.6
3, 0.4, 0.2, NA
4, ...
My idea is to use a paired T-test, which I am not sure about and leads me to the following 2 problems:
I would need two t-tests, one for comparing the means of A and B and one for B and C. So I would get also two p-values, but I would prefer to have everything in one test an get only one p value, if there is a possibility.
I have missing Values in my table, cause not every patient has all 3 sypmtoms. How do I handle them?
So my questions are if my idea (paired t-tests) is correct, how to handle the two issues above or if it is not correct. What would then be a better idea?