Let's say I have blood samples of whiteblood cells ($x$) and viruses ($v$). Space has been discretized in $LL$ spaces. They have a $p_v$ probability of interacting when found in the same space. I want to know at each sample how many $x$ and $v$ are interacting.
Here is a naive implementation of the model:
LL <- 1e4
nx <- 5000
nv <- 500
x <- c(rep(1,nx),rep(0, LL-nx))
v <- c(rep(1,nv),rep(0, LL-nv))
pv <- 0.8
t1 <- sapply(1:1e4, function(z) sum(sample(v)==1 & sample(x)==1 & runif(LL)<pv))
t1 represents how many times $x$ and $v$ interacted, in each of 1e4 samples.
I want to model this using a probability distribution, in order to make it more efficient. I think we can see this problem as a Binomial (and later maybe approximate with a Poisson), but I am not sure what arguments to pass to the Binomial. I can think of:
- at each space, there is a probability they are interacting, depending on their concentrations
- for each $x$, the probability is given by the concentration of $v$
- for each $v$, the probability is given by the concentration of $x$
In the first two cases, I have to limit the Binomial result to $nv$, which is the lowest value of molecules, and thus no more interactions than those are possible.
limit <- function(z)
sapply(z, function(z) min(z,nx,nv))
t3 <- limit(rbinom(1e4, LL, pv*nx*nv/(LL*LL)))
t4 <- limit(rbinom(1e4, max(nx,nv), pv*min(nx,nv)/LL))
t5 <- rbinom(1e4, min(nx,nv), pv*max(nx,nv)/LL)
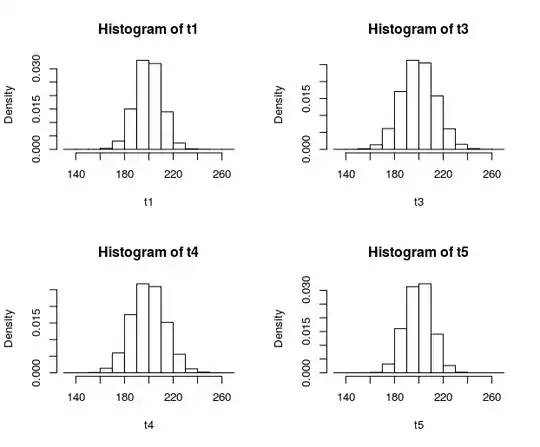
The histograms:
par(mfrow=c(2,3))
b <- seq(130, 270, 10)
hist(t1,b,FALSE)
hist(t3,b,FALSE)
hist(t4,b,FALSE)
hist(t5,b,FALSE)
print(sprintf("mean: %.3f x %.3f x %.3f x %.3f x %.3f",mean(t1),mean(t3),mean(t4),mean(t5)))
print(sprintf("sd: %.3f x %.3f x %.3f x %.3f x %.3f",sd(t1),sd(t3),sd(t4),sd(t5)))
Output:
[1] "mean: 200.036 x 200.281 x 199.991 x 200.108"
[1] "sd: 10.660 x 14.076 x 13.817 x 10.841"
The result suggests the latter maybe. Would love to have some feedback on this though. Thank you so much !