I am trying to draw a plot of the decision function ($f(x)=sign(wx+b)$ which can be obtain by fit$decision.values in R using the svm function of e1071 package) versus another arbitrary values.
From svm documentation, for binary classification the new sample can be classified based on the sign of f(x), so I can draw a vertical line on zero and the two classes can be separated from each other. I’ve used the example form here.
require(e1071)
# Subset the iris dataset to only 2 labels and 2 features
iris.part = subset(iris, Species != 'setosa')
iris.part$Species = factor(iris.part$Species)
iris.part = iris.part[, c(1,2,5)]
# Fit svm model
fit = svm(Species ~ ., data=iris.part, type='C-classification', kernel='linear')
> head(fit$decision.values)
versicolor/virginica
51 -1.3997066
52 -0.4402254
53 -1.1596819
54 1.7199970
55 -0.2796942
56 0.9996141
...
Tabulate actual class labels vs. model predictions:
> table(Actual=iris.part$Species, Fitted=pred)
Fitted
Actual versicolor virginica
versicolor 38 12
virginica 15 35
Plot of decision function
fit$decision.values
plot(fx,fy,pch=rep(c(3,1),c(50,50)),col=rep(1:2,c(50,50)))
abline(v=0)
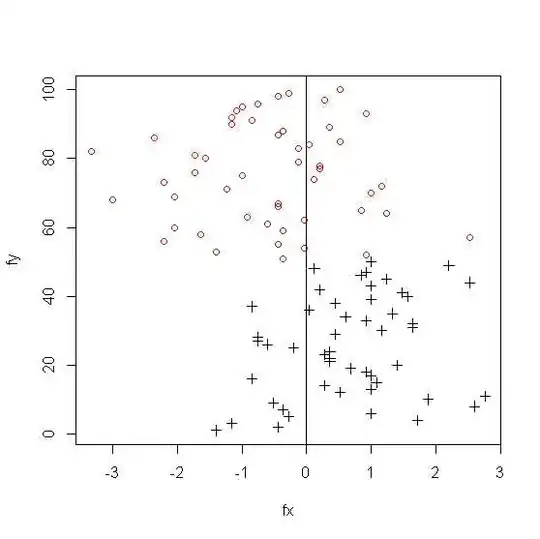
It can be seen that there is 15 and 12 misclassified example in class 1 and class 2 respectively.
The resulting plot for 3 class svm ;
But not sure how to deal with multi-class classification; can anyone help me on that? Is there any way I can draw boundary line that can separate $f(x) $ of each class from the others and shows the number of misclassified observation similar to the results of the following table?
>fit = svm(Species ~ ., data=iris, type='C-classification', kernel='linear')
>pred = predict(fit, iris)
Tabulate actual class labels vs. model predictions:
> table(Actual=iris$Species, Fitted=pred)
Fitted
Actual setosa versicolor virginica
setosa 50 0 0
versicolor 0 46 4
virginica 0 1 49