I have an obviously bimodal distribution of values, which I seek to fit. The data can be fit well with either 2 normal functions (bimodal) or with 3 normal functions. Additionally, there is a plausible physical reason for fitting the data with 3.
The more parameters that are introduced, the more perfect the fit will be, as with enough constants, one can "fit an elephant".
Here is the distribution, fit with the sum of 3 normal (Gaussian) curves:
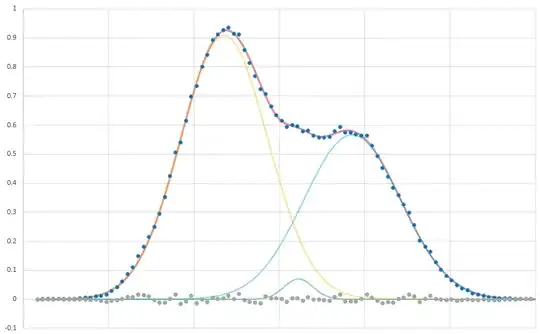
These are the data for each fit. I'm not sure what test I should be applying here to determine the fit. The data consists of 91 points.
1 Normal Function:
- RSS: 1.06231
- X^2: 3.1674
- F.Test: 0.3092
2 Normal Functions:
- RSS: 0.010939
- X^2: 0.053896
- F.Test: 0.97101
3 Normal Functions:
- RSS: 0.00536
- X^2: 0.02794
- F.Test: 0.99249
What is the correct statistical test that can be applied to determine which of these 3 fits is best? Obviously, the 1 normal function fit is inadequate. So how can I discriminate between 2 and 3?
To add, I'm mostly doing this with Excel and a little Python; I don't yet have familiarity with R or other statistical languages.