I want to find statistical criteria for signal registration (see pict). When I don't have a signal I registrate a simple gaussian two-dimenshial distribution. When I have a signal I have gaussian plus some "tail" (horizontal on the picture). Direction of a "tail" may differs, but I have some minimum length of a tail for a proper signal.
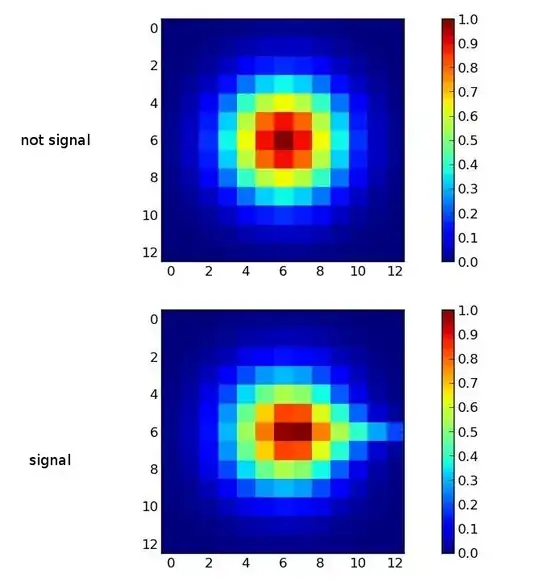
I tried to use chi square criteria but faild. p-value for my signal was 1.0. What kind of a criteria can I use in this case?
There is a sample data:
A signal:
[[ 0. 0. 0. 0.01 0.01 0.01 0.02 0.01 0.01 0.01 0. 0. 0. ]
[ 0. 0. 0.01 0.02 0.03 0.05 0.05 0.05 0.03 0.02 0.01 0. 0. ]
[ 0. 0.01 0.02 0.05 0.09 0.13 0.14 0.13 0.09 0.05 0.02 0.01 0. ]
[ 0.01 0.02 0.05 0.11 0.2 0.27 0.31 0.28 0.2 0.11 0.05 0.02 0.01]
[ 0.01 0.03 0.09 0.2 0.34 0.48 0.56 0.52 0.37 0.21 0.1 0.03 0.01]
[ 0.01 0.05 0.13 0.27 0.48 0.69 0.84 0.83 0.62 0.4 0.22 0.08 0.03]
[ 0.02 0.05 0.14 0.31 0.54 0.78 0.99 1. 0.77 0.56 0.4 0.29 0.19]
[ 0.01 0.05 0.13 0.27 0.48 0.69 0.84 0.83 0.62 0.4 0.22 0.08 0.03]
[ 0.01 0.03 0.09 0.2 0.34 0.48 0.56 0.52 0.37 0.21 0.1 0.03 0.01]
[ 0.01 0.02 0.05 0.11 0.2 0.27 0.31 0.28 0.2 0.11 0.05 0.02 0.01]
[ 0. 0.01 0.02 0.05 0.09 0.13 0.14 0.13 0.09 0.05 0.02 0.01 0. ]
[ 0. 0. 0.01 0.02 0.03 0.05 0.05 0.05 0.03 0.02 0.01 0. 0. ]
[ 0. 0. 0. 0.01 0.01 0.01 0.02 0.01 0.01 0.01 0. 0. 0. ]]
Not a signal:
[[ 0. 0. 0. 0.01 0.01 0.02 0.02 0.02 0.01 0.01 0. 0. 0. ]
[ 0. 0. 0.01 0.02 0.04 0.06 0.06 0.06 0.04 0.02 0.01 0. 0. ]
[ 0. 0.01 0.03 0.06 0.11 0.15 0.17 0.15 0.11 0.06 0.03 0.01 0. ]
[ 0.01 0.02 0.06 0.14 0.24 0.33 0.37 0.33 0.24 0.14 0.06 0.02 0.01]
[ 0.01 0.04 0.11 0.24 0.41 0.57 0.64 0.57 0.41 0.24 0.11 0.04 0.01]
[ 0.02 0.06 0.15 0.33 0.57 0.8 0.9 0.8 0.57 0.33 0.15 0.06 0.02]
[ 0.02 0.06 0.17 0.37 0.64 0.9 1. 0.9 0.64 0.37 0.17 0.06 0.02]
[ 0.02 0.06 0.15 0.33 0.57 0.8 0.9 0.8 0.57 0.33 0.15 0.06 0.02]
[ 0.01 0.04 0.11 0.24 0.41 0.57 0.64 0.57 0.41 0.24 0.11 0.04 0.01]
[ 0.01 0.02 0.06 0.14 0.24 0.33 0.37 0.33 0.24 0.14 0.06 0.02 0.01]
[ 0. 0.01 0.03 0.06 0.11 0.15 0.17 0.15 0.11 0.06 0.03 0.01 0. ]
[ 0. 0. 0.01 0.02 0.04 0.06 0.06 0.06 0.04 0.02 0.01 0. 0. ]
[ 0. 0. 0. 0.01 0.01 0.02 0.02 0.02 0.01 0.01 0. 0. 0. ]]