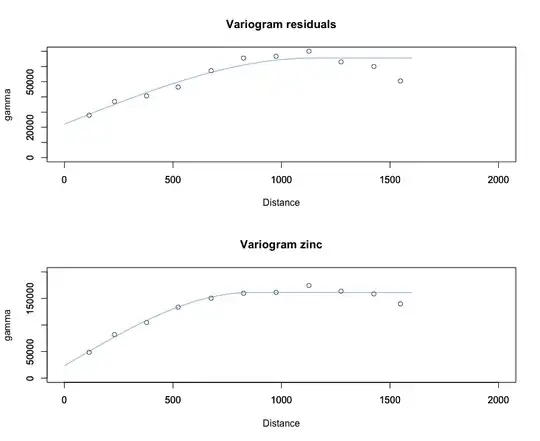
I am working with the build in dataset meuse, which has 155 measurements of Zinc and the distance to the river "Meuse".(http://rspatial.r-forge.r-project.org/gallery/).
Now I am trying to imitate universal kriging. In R, you first model a variogram, which is used to model the spatial correlation between the observations.
Is there a difference in doing the following:
- First make a variogram of the zinc measured values, and use universal kriging;
- First regress zinc on distance, and make a variogram with the residuals.
Is somebody able to explain the deeper thoughts about these two different approaches? Can this be linked with the difference between conditionally autoregressive and simultaneous autoregressive models?