I am using Silhouette width to compute the best value for k in k-means. As I am performing document clustering, I am calculating the values of a and b as follows in Python:
a = distance(data[index], centroids[clusters[index]], metric=metric, p=p)
b = min([distance(data[index], c) for i,c in enumerate(centroids) if i != currentindex])
score = float(b - a) / max(a, b) if max(a, b) > 0 else 0.0
The original formula from the Wikipedia page is the following:
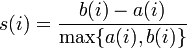
I am using a cosine similarity measure to compute the distances and was wondering if this formula needs to be changed or it can be left as is. In the above snippet of code, the function distance computes the cosine similarity metric.