[This question is related to 1 and 2 on this site.]
I fit a Cox model with these three time dependent variables: {s:numeric, C:binary, l:numeric }. I have 1069 events and around half of that in right censored observations.
model<- coxph( Surv(start, stop, event) ~ s+ C+ l, data=dat)
coef exp(coef) se(coef) z p
s 3.32 27.61 0.0825 40.2 0
CTRUE 1.36 3.88 0.1086 12.5 0
l 0.08 1.08 0.0077 10.4 0
Likelihood ratio test=990 on 3 df, p=0 n= 798431, number of events= 1069
I check the proportional hazards (PH) assumption for variables and the global model:
cox.zph(model)
rho chisq p
s 0.2066 12.536 0.000399
CTRUE 0.0453 2.212 0.136984
l 0.0461 0.835 0.360842
GLOBAL NA 14.684 0.002108
I can see that s violates the PH assumption, resulting in strong evidence of non-proportional hazards for the global model.
Also from the plots below, which show the estimated regression coefficients as functions of time, I see that the coefficients for s looks suspicious, especially the section I circled in red:
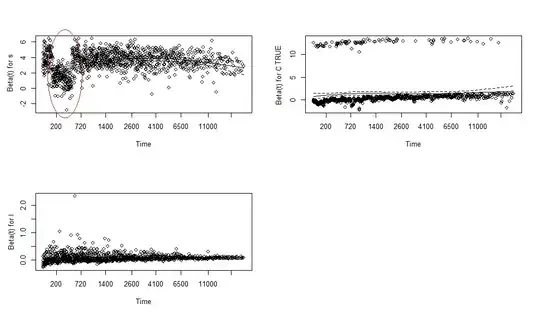
To treat this, I add a time interaction with s and refit the model. However, if you look at these new results, the p value for the s:stop coefficient is higher than my cut-off p= 0.05, so not statistically significant(!) :
model2<- coxph( Surv(start, stop, event) ~ s+ s:stop + C+ l, data=dat)
coef exp(coef) se(coef) z p
s 3.25e+00 25.84 9.32e-02 34.89 0.000
CTRUE 1.36e+00 3.88 1.09e-01 12.49 0.000
l 7.98e-02 1.08 7.72e-03 10.34 0.000
s:stop 4.42e-05 1.00 2.63e-05 1.68 0.094
Likelihood ratio test=993 on 4 df, p=0 n= 798431, number of events= 1069
Question 1: Can someone help me interpret the Schoenfeld graphs I plotted, especially what the red region in the s plot menas?
Question 2: Any general ideas on what is going on here as far as s is concerned?
P.S: I am reading the paper* below in the mean time to get a better insight but any answers by the more experienced will be very valuable.
*Patricia M Grambsch and Terry M Therneau."Proportional hazard tests and diagnostics based on weighted residuals", Biometrika, 81:515-526, 1994.
EDIT:
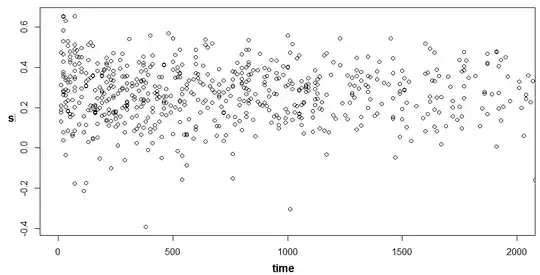
As per @EdM's comment, I plotted the s values per event time, zooming on in between the time=150 - time=700 region. Nothing particularly suspicious. So the actual s values seem normal but the coefficients the fit gives them are rather strange.