I am currently experimenting with PYMC and I am trying out a simple example so that I start learning how things work (I am also a Python beginner, but an experienced machine learner).
I have set myself the following very simple model selection task:
Imagine we have a dataset that has been entirely generated either from model 0 or model 1 (not a mixture).
Model 0 is a normal distribution with mean m0 and standard deviation s0.
Likewise, model 1 is also a normal distribution with mean m1 and standard deviation s1.
We assume that we know all parameters m0,s0,m1,s1.
We postulate a hidden binary random variable V which indicates the model that really generated the dataset.
What we would like to do is infer the posterior of V.
I have made an attempt in the code below, but unfortunately after trying for a couple of days, I capitulate and throw myself at your feet for assistance.
The example is of course very simple and may appear contrived. What I would like to do in the future, is use PYMC for model selection. Hence, how one correctly handles a hidden indicator variable like V is important to me. Unfortunately, in my code I keep getting weird results for the posterior of V.
I hope the code below makes sense. I thank you all for your time.
from pymc import *
import matplotlib
from pylab import hist, show, plot, title, figure
import numpy as np
from numpy import *
#-----------------------------------------------
# Generate some data
#-----------------------------------------------
# Define means for gaussians
mu0 = 5.0
mu1 = -5.0
# Define standard deviations for gaussians
s0 = 1.0
s1 = 1.0
# This variable chooses from which model we generate the observed data.
# It can be either 0 or 1
true_model = 0
# number of data items
numData = 100
if true_model==0:
# Generate data from first model
data = np.random.randn(numData,1)*s0 + mu0
elif true_model==1:
# Generate data from second model
data = np.random.randn(numData,1)*s1 + mu1
#-----------------------------------------------
# Define variables
#-----------------------------------------------
PR = Dirichlet("priorOnModels", theta=(0.5,0.5)) # both models are equally likely
V = Categorical('V', p=PR)
#-----------------------------------------------
# Define model log-likelihood
#-----------------------------------------------
@potential
def mymodel_loglikel(sw=V):
if sw==0:
loglikel = distributions.normal_like(data, mu0, 1.0/s0**2)
elif sw==1:
loglikel = distributions.normal_like(data, mu1, 1.0/s1**2)
return loglikel
#-----------------------------------------------
# Run sampler
#-----------------------------------------------
simplemodel = Model([mymodel_loglikel, PR, V])
mc = MCMC(simplemodel)
mc.sample(iter=12000,burn=2000)
figure()
hist(data)
Matplot.plot(mc)
print "expectation of V variable is %.3f" % np.mean(mc.trace('V')[:])
show()
EDIT 2: Updated after Tom's suggestions
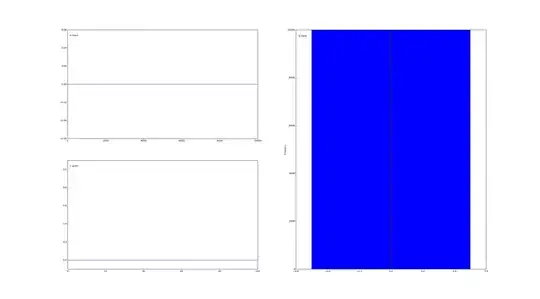
In response to a comment below, the weird behaviour I get concerning V, is that its mean posterior is always 0.0. Judging from the trace, it seems that V is not properly sampled. I suspect a minor programming error somewhere, but can't pinpoint it.
I attach below a figure of the trace plots of V.