The variable of interest is multinomially distributed with class (cell) probabilities: $p_1, p_2, ..., p_{10}$. Further, the classes are endowed with a natural order.
First attempt: smallest "predictive interval" containing $90\%$
p = [p1, ..., p10] # empirical proportions summing to 1
l = 1
u = length(p)
cover = 0.9
pmass = sum(p)
while (pmass - p[l] >= cover) OR (pmass - p[u] >= cover)
if p[l] <= p[u]
pmass = pmass - p[l]
l = l + 1
else # p[l] > p[u]
pmass = pmass - p[u]
u = u - 1
end
end
A non-parametric measure of the uncertainty (e.g., variance, confidence) in the $l,u$-quantile estimates could indeed be obtained by standard bootstrap methods.
Second approach: direct "bootstrap search"
Below I provide runable Matlab code that approaches the question directly from a bootstrap perspective (the code is not optimally vectorized).
%% set DGP parameters:
p = [0.35, 0.8, 3.5, 2.2, 0.3, 2.9, 4.3, 2.1, 0.4, 0.2];
p = p./sum(p); % true probabilities
ncat = numel(p);
cats = 1:ncat;
% draw a sample:
rng(1703) % set seed
nsamp = 10^3;
samp = datasample(1:10, nsamp, 'Weights', p, 'Replace', true);
Check that this makes sense.
psamp = mean(bsxfun(@eq, samp', cats)); % sample probabilities
bar([p(:), psamp(:)])
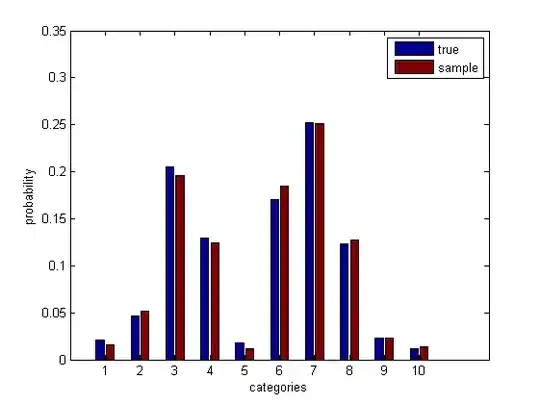
Run the bootstrap simulation.
%% bootstrap simulation:
rng(240947)
nboots = 2*10^3;
cover = 0.9;
conf = 0.95;
tic
Pmat = nan(nboots, ncat, ncat);
for b = 1:nboots
boot = datasample(samp, nsamp, 'Replace', true); % draw bootstrap sample
pboot = mean(bsxfun(@eq, boot', cats));
for l = 1:ncat
for u = l:ncat
Pmat(b, l, u) = sum(pboot(l:u));
end
end
end
toc % Elapsed time is 0.442703 seconds.
Filter from each bootstrap replicate the intervals, $[l,u]$, that contain at least $90\%$ probability mass and calculate a (frequentist) confidence estimate of those intervals.
conf_mat = squeeze(mean(Pmat >= cover, 1))
0 0 0 0 0 0 0 1.0000 1.0000 1.0000
0 0 0 0 0 0 0 1.0000 1.0000 1.0000
0 0 0 0 0 0 0 0.3360 0.9770 1.0000
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
Select those that satisfy the confidence desideratum.
[L, U] = find(conf_mat >= conf);
[L, U]
1 8
2 8
1 9
2 9
3 9
1 10
2 10
3 10
Convincing yourself that the above bootstrap method is valid
Bootstrap samples are intended to be stand-ins for something we would like to have, but do not, i.e.: new, independent draws from the true underlying population (short: new data).
In the example that I gave, we know the data generating process (DGP), therefore we could "cheat" and replace the code lines pertaining to bootstrap re-samples by new, independent draws from the actual DGP.
newsamp = datasample(cats, nsamp, 'Weights', p, 'Replace', true);
pnew = mean(bsxfun(@eq, newsamp', cats));
Then we can validate the bootstrap approach by comparing it to the ideal. Below are the results.
The confidence matrix from new, independent data draws:
0 0 0 0 0 0 0 1.0000 1.0000 1.0000
0 0 0 0 0 0 0 1.0000 1.0000 1.0000
0 0 0 0 0 0 0 0.4075 0.9925 1.0000
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
The corresponding $95\%$-confidence lower and upper bounds:
1 8
2 8
1 9
2 9
3 9
1 10
2 10
3 10
We find that the confidence matrices closely agree and that the bounds are identical... Thus validating the bootstrap approach.
