This question seems related, but the consensus was that the issue had to do scaling the data, which I do prior to training, so I don't think that's the issue:
I've uploaded a sample data set, and here is how I generated my model:
library(randomForest)
library(caret)
library(ggplot2)
data <- read.csv("http://pastebin.com/raw.php?i=mE5JL1dm")
data_pred <- data[, 1:(ncol(data) - 1)]
data_resp <- as.factor(data$y)
data_trans <- preProcess(data_pred, method = c("center", "scale"))
data_pred_scale <- predict(data_trans, data_pred)
trControl <- trainControl(method = "LGOCV", p = 0.9, savePredictions = T)
set.seed(123)
model <- train(x = data_pred_scale, y = data_resp,
method = "rf", scale = F,
trControl = trControl)
Here's what caret() reports as the model performance:
> model
Random Forest
516 samples
11 predictors
5 classes: '0', '0.5', '1', '1.5', '2'
No pre-processing
Resampling: Repeated Train/Test Splits Estimated (25 reps, 0.9%)
Summary of sample sizes: 468, 468, 468, 468, 468, 468, ...
Resampling results across tuning parameters:
mtry Accuracy Kappa Accuracy SD Kappa SD
2 0.747 0.663 0.0643 0.0853
6 0.76 0.68 0.0507 0.068
11 0.758 0.678 0.0574 0.0763
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was mtry = 6.
In my "real" model, I have a training/hold-out set, and creating two sets of plots showing model predictions for the training/hold-out sets vs. the corresponding true observations. That's when I noticed something that seemed odd to me.
# data set of model predictions on training data vs. actual observations
results <- data.frame(pred = predict(model, data_pred_scale),
obs = data_resp)
table(results)
obs
pred 0 0.5 1 1.5 2
0 148 0 0 0 0
0.5 0 132 0 0 0
1 0 0 139 0 0
1.5 0 0 0 38 0
2 0 0 0 0 59
And here's a plot confirming 100% accuracy on the training set:
p <- ggplot(results, aes(x = pred, y = obs))
p <- p + geom_jitter(position = position_jitter(width = 0.25, height = 0.25))
p
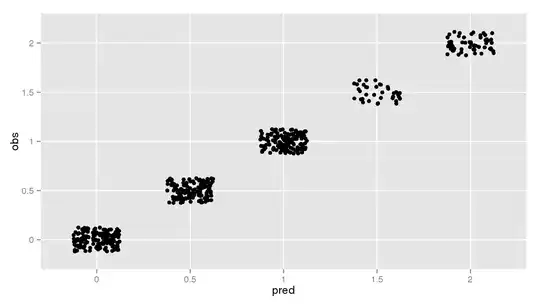
But if I look at the saved tuning predictions, subsetting only those where mtry = 6 (what caret reports as the final model), I don't get anywhere near that performance:
model_resamples <- model$pred[model$pred$mtry == 6, c("pred", "obs")
table(model_resamples)
0 0.5 1 1.5 2
0 296 69 5 0 0
0.5 51 228 48 0 0
1 3 28 255 24 9
1.5 0 0 16 32 15
2 0 0 1 19 101
And the same sort of plot:
p <- ggplot(model_resamples, aes(x = pred, y = obs))
p <- p + geom_jitter(position = position_jitter(width = 0.25, height = 0.25))
p
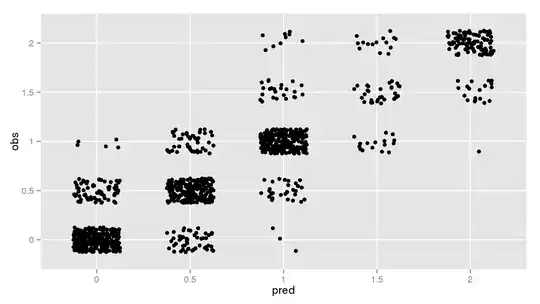
Is this just a case of over-fitting where holding out 10% of the data can create a 25% decrease in performance, yet the final model trained with the same parameters but all rows can yield 100% performance? It seemed unlikely, but that's the only thing coming to mind at the moment.
I just want to make sure there's nothing wrong in my training or predicting methods where I'm creating a problem where there shouldn't be one.
Note: I created the tables/plots prior to adding set.seed() in the model training code above. The exact table and plot may differ slightly, but in re-running, they general result is the same (perfect re-prediction vs. ~77% reported by the model). It didn't seem to warrant re-doing the results/plots above, so I left them.